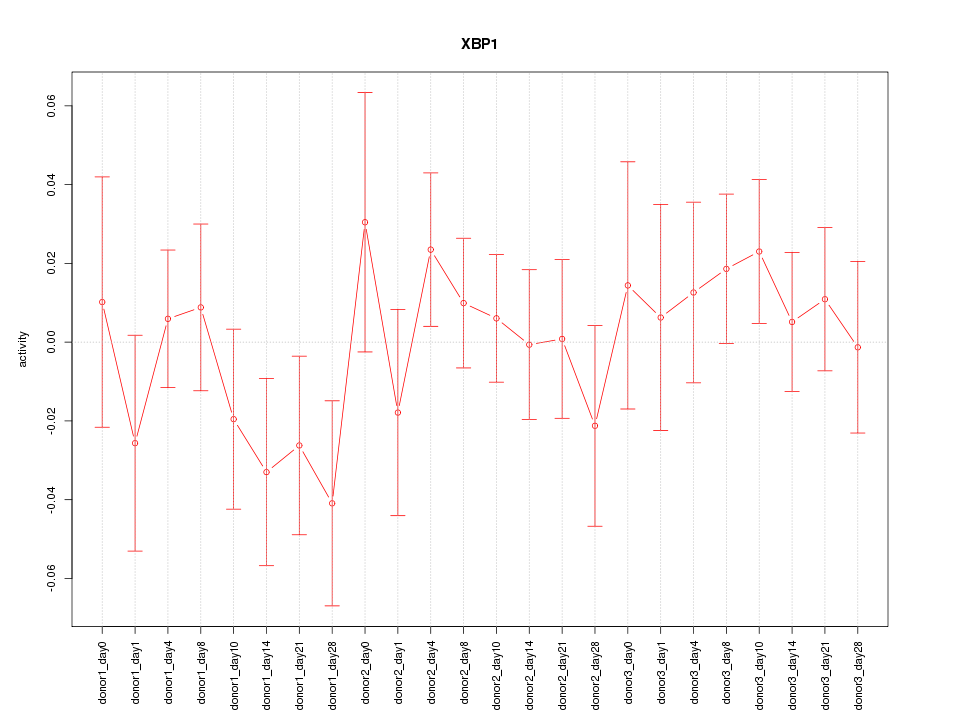
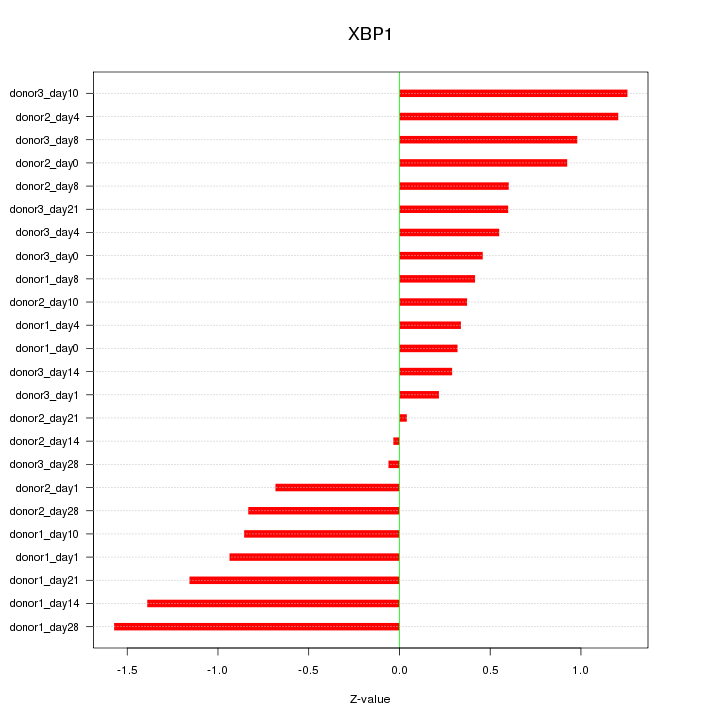
Motif ID: XBP1
Z-value: 0.797
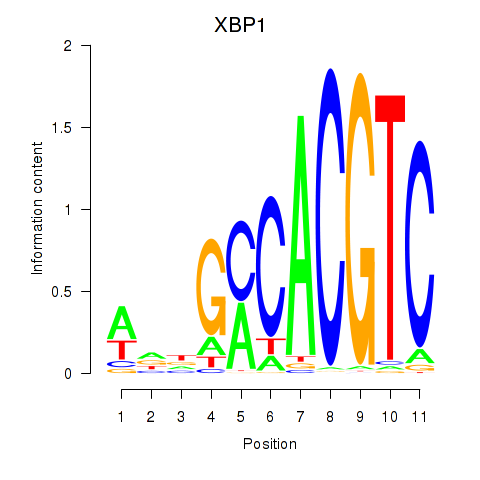
Transcription factors associated with XBP1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| XBP1 | ENSG00000100219.12 | XBP1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| XBP1 | hg19_v2_chr22_-_29196546_29196585, hg19_v2_chr22_-_29196030_29196121 | -0.14 | 5.3e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 1.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 1.0 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 0.5 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.1 | 0.7 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.6 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.1 | 0.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.9 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 | 0.4 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.1 | 0.4 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.8 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 1.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 0.7 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.3 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 1.0 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 1.0 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 0.2 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.1 | 0.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.3 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.1 | 0.9 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 | 0.6 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 1.9 | GO:0002076 | osteoblast development(GO:0002076) negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 0.9 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.5 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.7 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 1.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.3 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.3 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 1.3 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.4 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:1990737 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.3 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.6 | GO:0060707 | dorsal/ventral axis specification(GO:0009950) trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 1.1 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0002461 | tolerance induction dependent upon immune response(GO:0002461) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0097212 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) positive regulation of late endosome to lysosome transport(GO:1902824) positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 1.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 1.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.0 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.3 | 1.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 0.9 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.2 | 1.0 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 0.5 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 1.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.6 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.5 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 1.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.7 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.3 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 2.0 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 0.9 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.2 | 0.8 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 0.7 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.2 | 0.8 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.2 | 1.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.6 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.1 | 0.4 | GO:0008478 | pyridoxal kinase activity(GO:0008478) lithium ion binding(GO:0031403) |
| 0.1 | 0.7 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.6 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.3 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 0.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.3 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.1 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.5 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 0.3 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 0.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.3 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.9 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 1.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.0 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.7 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.1 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.0 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.6 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.8 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.4 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.3 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.9 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME_NUCLEOTIDE_LIKE_PURINERGIC_RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.7 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME_G_ALPHA1213_SIGNALLING_EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.6 | REACTOME_POST_TRANSLATIONAL_MODIFICATION_SYNTHESIS_OF_GPI_ANCHORED_PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.7 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME_RNA_POL_I_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 2.6 | REACTOME_SRP_DEPENDENT_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.2 | REACTOME_ADENYLATE_CYCLASE_INHIBITORY_PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 1.5 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |