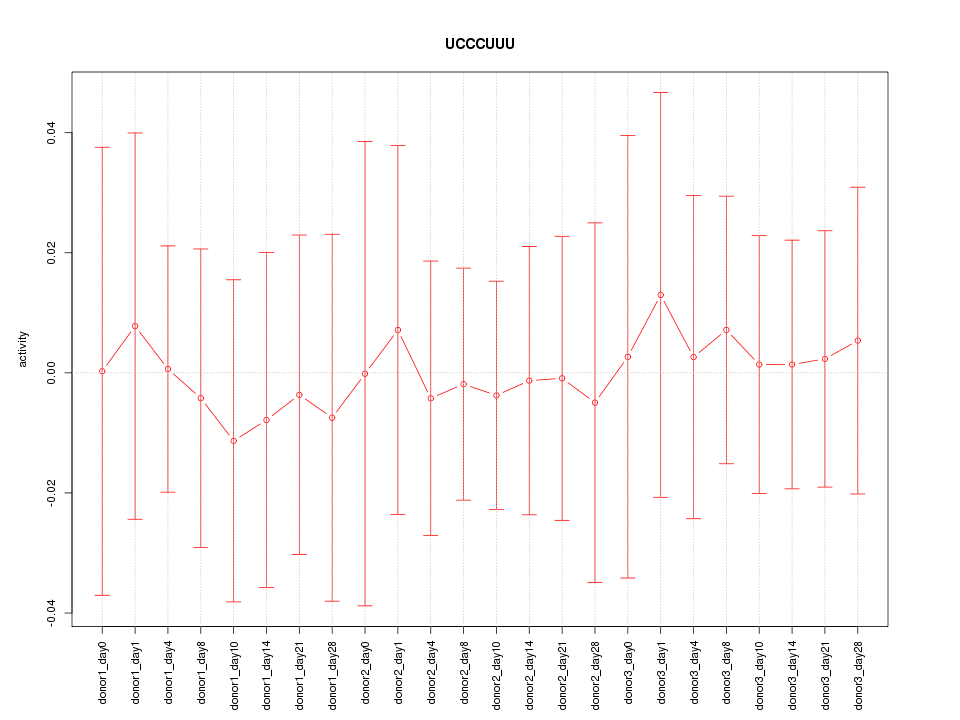
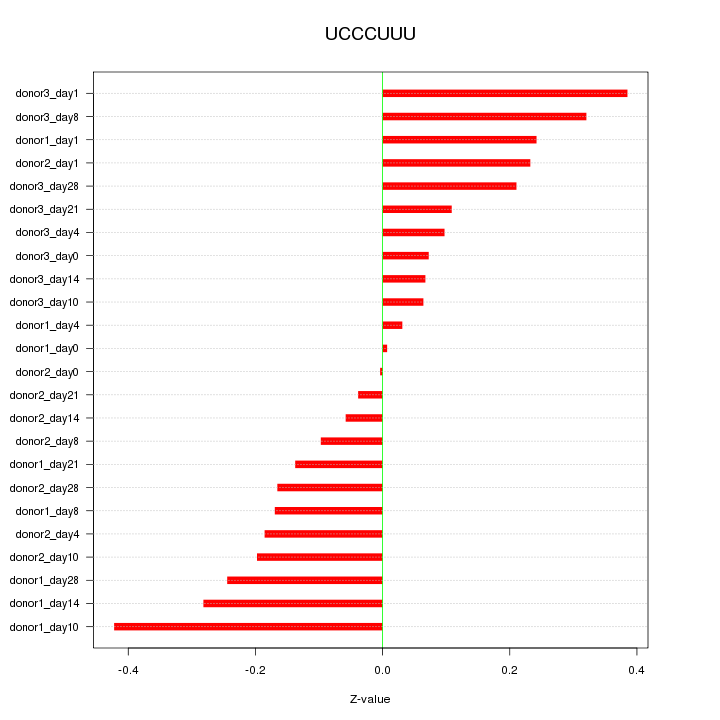
| 0.1 |
0.2 |
GO:0031052 |
mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.0 |
0.1 |
GO:0002276 |
basophil activation involved in immune response(GO:0002276) |
| 0.0 |
0.1 |
GO:0003358 |
noradrenergic neuron development(GO:0003358) |
| 0.0 |
0.1 |
GO:0042853 |
L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.0 |
0.1 |
GO:0003133 |
BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 |
0.1 |
GO:0097051 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 |
0.2 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.1 |
GO:0099590 |
neurotransmitter receptor internalization(GO:0099590) |
| 0.0 |
0.1 |
GO:0061386 |
closure of optic fissure(GO:0061386) |
| 0.0 |
0.1 |
GO:0002514 |
B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 |
0.4 |
GO:0071481 |
cellular response to X-ray(GO:0071481) |
| 0.0 |
0.1 |
GO:0045226 |
extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 |
0.1 |
GO:0071109 |
superior temporal gyrus development(GO:0071109) |
| 0.0 |
0.1 |
GO:1902728 |
mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 |
0.1 |
GO:0036510 |
trimming of terminal mannose on C branch(GO:0036510) |
| 0.0 |
0.0 |
GO:0002384 |
hepatic immune response(GO:0002384) |
| 0.0 |
0.1 |
GO:0035583 |
sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 |
0.1 |
GO:1902954 |
regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.0 |
0.1 |
GO:0071477 |
hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 |
0.1 |
GO:1902044 |
regulation of Fas signaling pathway(GO:1902044) |
| 0.0 |
0.0 |
GO:1902534 |
single-organism membrane invagination(GO:1902534) |
| 0.0 |
0.0 |
GO:0035281 |
pre-miRNA export from nucleus(GO:0035281) |
| 0.0 |
0.0 |
GO:0032484 |
Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 |
0.1 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.0 |
0.0 |
GO:1901873 |
regulation of post-translational protein modification(GO:1901873) |
| 0.0 |
0.1 |
GO:0061669 |
spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |