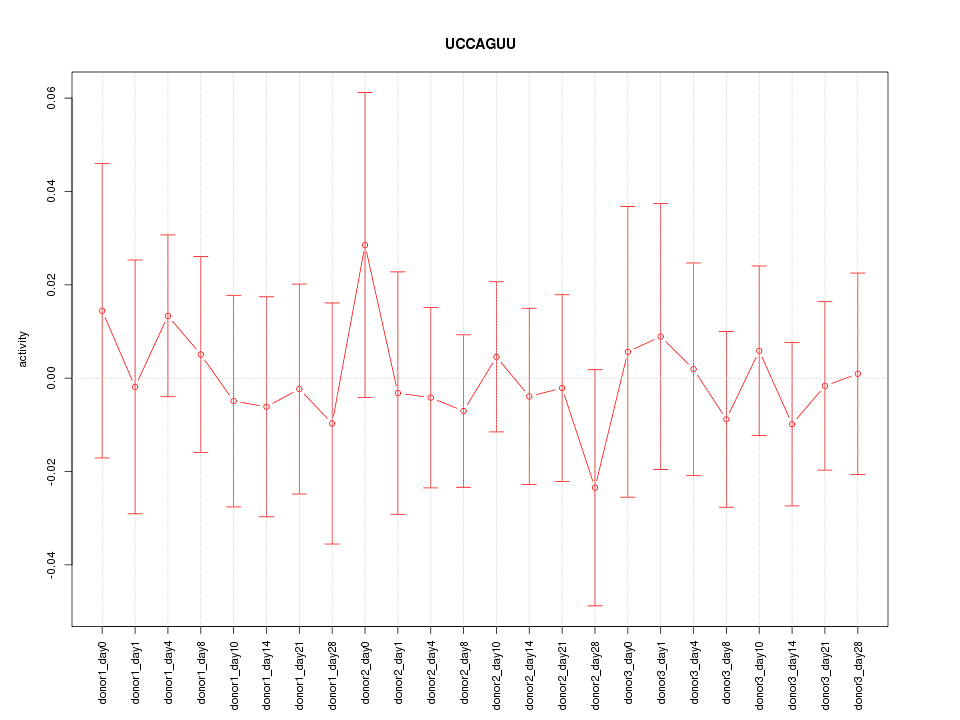
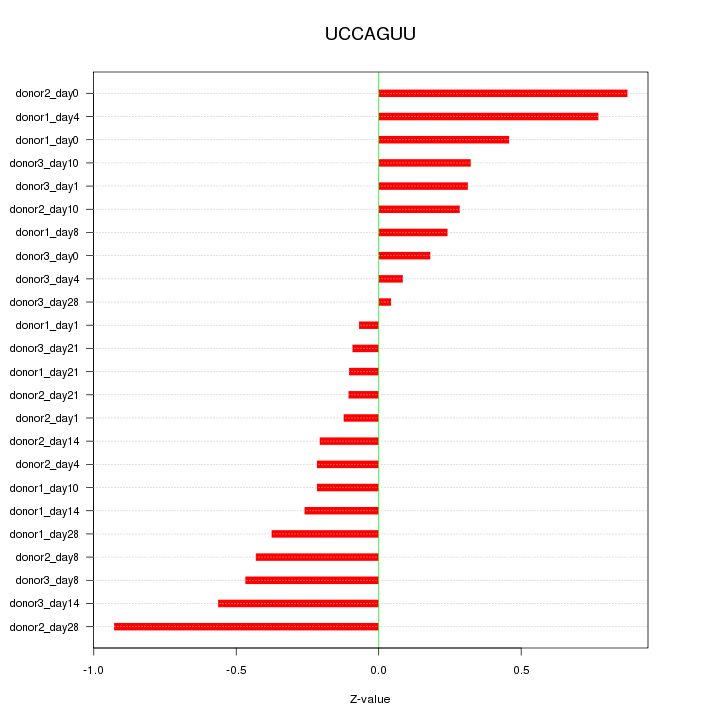
| 0.4 |
1.1 |
GO:2000097 |
chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.3 |
1.0 |
GO:0060516 |
primary prostatic bud elongation(GO:0060516) |
| 0.2 |
1.2 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.2 |
0.6 |
GO:0015920 |
lipopolysaccharide transport(GO:0015920) |
| 0.1 |
0.6 |
GO:0048880 |
sensory system development(GO:0048880) |
| 0.1 |
0.4 |
GO:0046909 |
intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 |
0.4 |
GO:0035625 |
epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.1 |
0.5 |
GO:0031585 |
regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) secretory granule localization(GO:0032252) endoplasmic reticulum localization(GO:0051643) |
| 0.1 |
1.2 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 |
0.2 |
GO:0014740 |
negative regulation of muscle hyperplasia(GO:0014740) |
| 0.1 |
0.4 |
GO:0061767 |
negative regulation of lung blood pressure(GO:0061767) |
| 0.1 |
0.4 |
GO:0071477 |
hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 |
0.2 |
GO:0060435 |
bronchiole development(GO:0060435) |
| 0.1 |
0.2 |
GO:0021816 |
lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.1 |
0.2 |
GO:2001045 |
negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 |
0.4 |
GO:0006065 |
UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 |
0.3 |
GO:0001712 |
ectodermal cell fate commitment(GO:0001712) |
| 0.1 |
0.2 |
GO:0070563 |
negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 |
0.1 |
GO:1900239 |
phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.1 |
0.2 |
GO:0061073 |
ciliary body morphogenesis(GO:0061073) |
| 0.0 |
0.3 |
GO:0032185 |
septin cytoskeleton organization(GO:0032185) |
| 0.0 |
0.2 |
GO:0033078 |
extrathymic T cell differentiation(GO:0033078) |
| 0.0 |
0.3 |
GO:0071461 |
cellular response to redox state(GO:0071461) |
| 0.0 |
0.1 |
GO:0060490 |
orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 |
0.2 |
GO:0035672 |
oligopeptide transmembrane transport(GO:0035672) |
| 0.0 |
0.1 |
GO:1904760 |
myofibroblast differentiation(GO:0036446) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 |
0.8 |
GO:0071481 |
cellular response to X-ray(GO:0071481) |
| 0.0 |
0.3 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.0 |
0.2 |
GO:0001927 |
exocyst assembly(GO:0001927) |
| 0.0 |
0.2 |
GO:0055064 |
chloride ion homeostasis(GO:0055064) |
| 0.0 |
0.1 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 |
0.3 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.0 |
0.2 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.0 |
0.1 |
GO:0052047 |
interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 |
0.4 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 |
0.1 |
GO:0071484 |
cellular response to light intensity(GO:0071484) |
| 0.0 |
0.2 |
GO:1904627 |
response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 |
0.2 |
GO:0007185 |
transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 |
0.3 |
GO:0060591 |
chondroblast differentiation(GO:0060591) |
| 0.0 |
0.3 |
GO:0060174 |
limb bud formation(GO:0060174) |
| 0.0 |
0.4 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.0 |
0.2 |
GO:0030578 |
PML body organization(GO:0030578) |
| 0.0 |
0.3 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 |
0.1 |
GO:0015917 |
aminophospholipid transport(GO:0015917) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 |
0.4 |
GO:0030043 |
actin filament fragmentation(GO:0030043) |
| 0.0 |
0.1 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.1 |
GO:0032455 |
nerve growth factor processing(GO:0032455) |
| 0.0 |
0.1 |
GO:0038109 |
response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 |
0.1 |
GO:1903691 |
positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 |
0.4 |
GO:0060707 |
trophoblast giant cell differentiation(GO:0060707) |
| 0.0 |
0.1 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 |
0.4 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.1 |
GO:0071409 |
cellular response to cycloheximide(GO:0071409) |
| 0.0 |
0.1 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.0 |
0.2 |
GO:0000727 |
double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 |
0.1 |
GO:1903551 |
regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 |
0.1 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.0 |
0.2 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) cartilage morphogenesis(GO:0060536) |
| 0.0 |
0.1 |
GO:1905205 |
positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 |
0.1 |
GO:0071947 |
protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 |
0.3 |
GO:2000587 |
negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 |
0.0 |
GO:0009826 |
unidimensional cell growth(GO:0009826) |
| 0.0 |
0.1 |
GO:0044791 |
modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 |
0.1 |
GO:0032000 |
positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 |
0.1 |
GO:0042816 |
vitamin B6 metabolic process(GO:0042816) |
| 0.0 |
0.1 |
GO:0001880 |
Mullerian duct regression(GO:0001880) |
| 0.0 |
0.1 |
GO:0010816 |
substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 |
0.1 |
GO:0021769 |
orbitofrontal cortex development(GO:0021769) |
| 0.0 |
0.1 |
GO:0036289 |
peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 |
0.4 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 |
0.1 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.0 |
0.2 |
GO:1901725 |
regulation of histone deacetylase activity(GO:1901725) |
| 0.0 |
0.1 |
GO:0010606 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 |
0.1 |
GO:0032417 |
positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 |
0.1 |
GO:0032287 |
peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 |
0.2 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 |
0.1 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 |
0.0 |
GO:0019521 |
pentose-phosphate shunt, oxidative branch(GO:0009051) aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 |
0.2 |
GO:0036010 |
protein localization to endosome(GO:0036010) |
| 0.0 |
0.1 |
GO:0001675 |
acrosome assembly(GO:0001675) |
| 0.0 |
0.0 |
GO:0006663 |
platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 |
0.0 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.0 |
0.1 |
GO:0023019 |
signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 |
0.1 |
GO:0045086 |
positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 |
0.1 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |