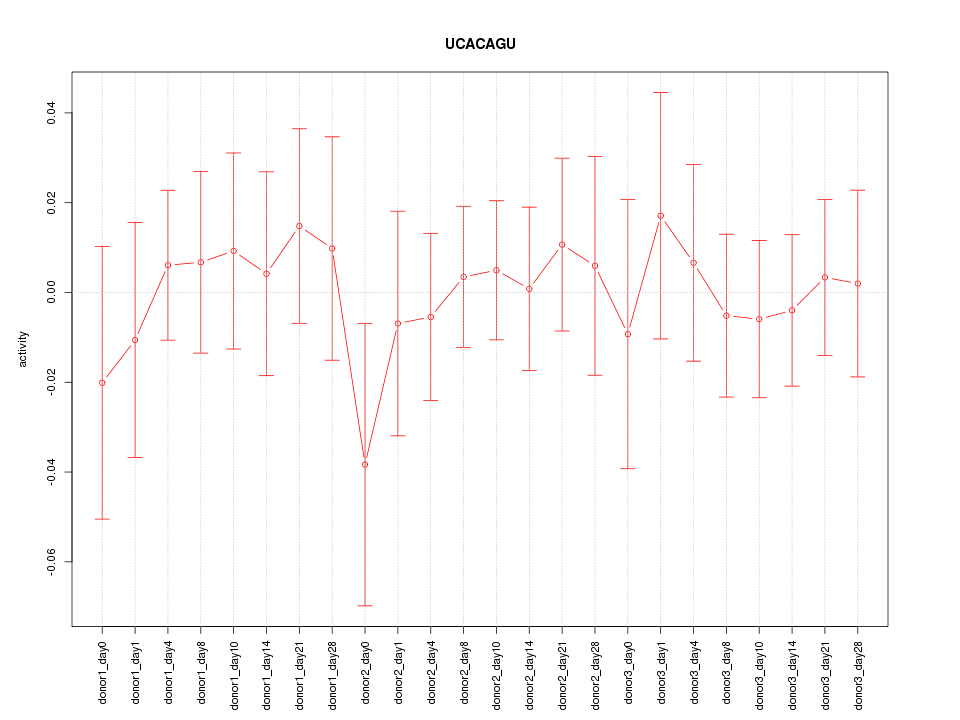
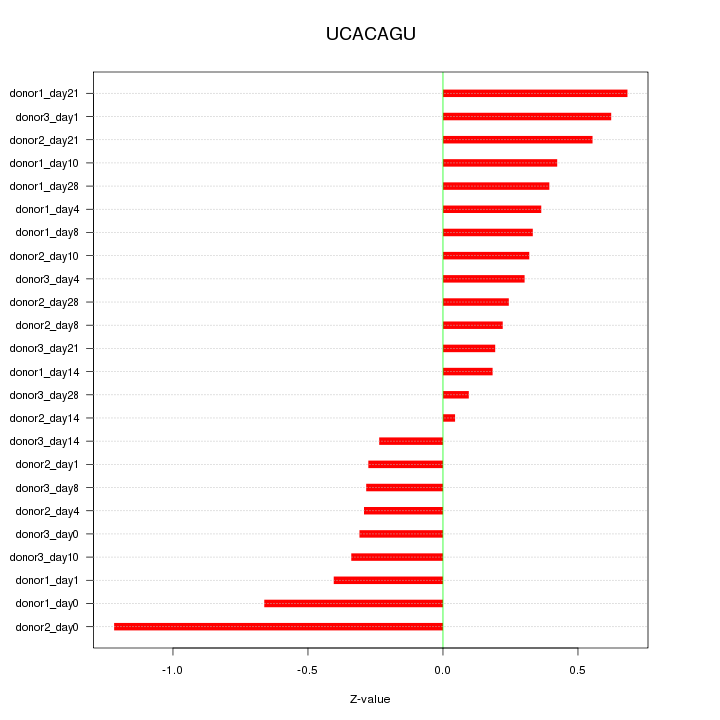
| 0.2 |
0.2 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.2 |
0.6 |
GO:0021524 |
visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) |
| 0.2 |
1.1 |
GO:0008063 |
Toll signaling pathway(GO:0008063) |
| 0.2 |
0.5 |
GO:0002071 |
glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.1 |
1.0 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 |
0.6 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 |
0.4 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.1 |
0.6 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) seminal vesicle development(GO:0061107) |
| 0.1 |
0.4 |
GO:1990535 |
transformation of host cell by virus(GO:0019087) neuron projection maintenance(GO:1990535) |
| 0.1 |
0.7 |
GO:1900020 |
regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 |
0.3 |
GO:0045976 |
negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.1 |
0.3 |
GO:1900169 |
regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 |
0.3 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 |
0.3 |
GO:2000276 |
negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 |
0.4 |
GO:0099590 |
neurotransmitter receptor internalization(GO:0099590) |
| 0.1 |
0.5 |
GO:0060373 |
regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 |
1.1 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.1 |
0.2 |
GO:2001245 |
regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 |
0.2 |
GO:0033024 |
mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.1 |
0.4 |
GO:0048861 |
leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 |
0.2 |
GO:0097114 |
NMDA glutamate receptor clustering(GO:0097114) |
| 0.1 |
0.3 |
GO:0061304 |
retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 |
0.6 |
GO:0018103 |
protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 |
0.2 |
GO:0045082 |
positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.1 |
0.2 |
GO:1900039 |
positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 |
0.2 |
GO:1905246 |
regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 |
0.4 |
GO:1901341 |
activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 |
0.2 |
GO:0014054 |
positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.1 |
0.3 |
GO:0045829 |
negative regulation of isotype switching(GO:0045829) |
| 0.1 |
0.3 |
GO:1905098 |
negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 |
0.2 |
GO:0001893 |
maternal placenta development(GO:0001893) |
| 0.1 |
0.3 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.1 |
0.4 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.1 |
0.5 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 |
0.2 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 0.1 |
0.3 |
GO:2000809 |
positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 |
0.4 |
GO:1990034 |
cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 |
0.7 |
GO:0061000 |
negative regulation of dendritic spine development(GO:0061000) |
| 0.1 |
0.6 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.1 |
0.2 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.0 |
0.3 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.0 |
0.2 |
GO:0018242 |
protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 |
0.1 |
GO:0034136 |
negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) |
| 0.0 |
0.3 |
GO:0021831 |
embryonic olfactory bulb interneuron precursor migration(GO:0021831) |
| 0.0 |
0.2 |
GO:0060021 |
palate development(GO:0060021) |
| 0.0 |
0.2 |
GO:0032849 |
positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 |
0.2 |
GO:0072658 |
maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 |
0.3 |
GO:2000323 |
negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 |
0.1 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.0 |
0.4 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.0 |
0.0 |
GO:2001038 |
regulation of cellular response to drug(GO:2001038) |
| 0.0 |
0.3 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.0 |
0.2 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 |
0.6 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.0 |
0.3 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 |
0.1 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.0 |
0.4 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.0 |
0.6 |
GO:0019388 |
galactose catabolic process(GO:0019388) |
| 0.0 |
0.2 |
GO:1904953 |
Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904953) |
| 0.0 |
0.1 |
GO:0048372 |
lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) |
| 0.0 |
0.1 |
GO:1902725 |
negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 |
0.1 |
GO:0098582 |
innate vocalization behavior(GO:0098582) |
| 0.0 |
0.2 |
GO:0033183 |
negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 |
0.1 |
GO:0044691 |
tooth eruption(GO:0044691) |
| 0.0 |
0.1 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 |
0.6 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.2 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.0 |
0.1 |
GO:0032240 |
negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 |
0.3 |
GO:0060579 |
ventral spinal cord interneuron fate commitment(GO:0060579) cell fate commitment involved in pattern specification(GO:0060581) |
| 0.0 |
0.3 |
GO:0072383 |
plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 |
0.1 |
GO:0060721 |
spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 |
0.2 |
GO:0046604 |
positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 |
0.1 |
GO:1900224 |
positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 |
0.3 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 |
0.1 |
GO:1900222 |
regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 |
0.5 |
GO:0043584 |
nose development(GO:0043584) |
| 0.0 |
0.2 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.0 |
1.2 |
GO:0006198 |
cAMP catabolic process(GO:0006198) |
| 0.0 |
0.1 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.0 |
0.3 |
GO:0061088 |
regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 |
0.1 |
GO:0097070 |
ductus arteriosus closure(GO:0097070) |
| 0.0 |
0.1 |
GO:1902530 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 |
0.1 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.0 |
0.1 |
GO:0060012 |
synaptic transmission, glycinergic(GO:0060012) |
| 0.0 |
0.5 |
GO:1903204 |
negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 |
0.1 |
GO:0071072 |
negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 |
0.2 |
GO:0045876 |
positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 |
0.3 |
GO:0006048 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 |
0.3 |
GO:2000580 |
positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 |
0.2 |
GO:0060013 |
righting reflex(GO:0060013) |
| 0.0 |
0.3 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 |
0.1 |
GO:1902617 |
response to fluoride(GO:1902617) |
| 0.0 |
0.0 |
GO:0051533 |
positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 |
0.1 |
GO:1902809 |
regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 |
0.2 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.0 |
0.2 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.0 |
0.1 |
GO:0097360 |
chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 |
0.1 |
GO:0034670 |
chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 |
0.2 |
GO:0070236 |
negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 |
0.3 |
GO:0042340 |
keratan sulfate catabolic process(GO:0042340) |
| 0.0 |
0.1 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.2 |
GO:2000628 |
regulation of miRNA metabolic process(GO:2000628) |
| 0.0 |
0.1 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 |
0.0 |
GO:0021896 |
forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.0 |
0.1 |
GO:0035404 |
histone-serine phosphorylation(GO:0035404) |
| 0.0 |
0.0 |
GO:0010847 |
regulation of chromatin assembly(GO:0010847) |
| 0.0 |
0.1 |
GO:0031291 |
Ran protein signal transduction(GO:0031291) |
| 0.0 |
0.1 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 |
0.1 |
GO:0097026 |
dendritic cell dendrite assembly(GO:0097026) |
| 0.0 |
0.7 |
GO:2000144 |
positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 |
0.2 |
GO:1990416 |
cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 |
0.1 |
GO:0006679 |
glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 |
0.1 |
GO:1903371 |
regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 |
0.0 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 |
0.0 |
GO:2000270 |
negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 |
0.0 |
GO:0086100 |
endothelin receptor signaling pathway(GO:0086100) |
| 0.0 |
0.1 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.0 |
0.4 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.0 |
0.1 |
GO:1903588 |
negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 |
0.0 |
GO:0090291 |
negative regulation of osteoclast proliferation(GO:0090291) |
| 0.0 |
0.1 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 |
0.0 |
GO:0070681 |
glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 |
0.1 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.0 |
0.1 |
GO:0070245 |
positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 |
0.2 |
GO:0072189 |
ureter development(GO:0072189) |
| 0.0 |
0.2 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 |
0.1 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.2 |
GO:0019919 |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 |
0.0 |
GO:0014042 |
positive regulation of neuron maturation(GO:0014042) |
| 0.0 |
0.0 |
GO:0008355 |
olfactory learning(GO:0008355) |
| 0.0 |
0.2 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.0 |
0.1 |
GO:0033182 |
regulation of histone ubiquitination(GO:0033182) |
| 0.0 |
0.2 |
GO:0021694 |
cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 |
0.0 |
GO:0007497 |
posterior midgut development(GO:0007497) |
| 0.0 |
0.1 |
GO:0072332 |
intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 |
0.0 |
GO:0098907 |
protein localization to M-band(GO:0036309) regulation of SA node cell action potential(GO:0098907) |
| 0.0 |
0.2 |
GO:0021516 |
dorsal spinal cord development(GO:0021516) |
| 0.0 |
0.3 |
GO:0097094 |
craniofacial suture morphogenesis(GO:0097094) |
| 0.0 |
0.2 |
GO:0001682 |
tRNA 5'-leader removal(GO:0001682) |
| 0.0 |
0.1 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.0 |
0.1 |
GO:0002043 |
blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 |
0.1 |
GO:0043392 |
negative regulation of DNA binding(GO:0043392) |
| 0.0 |
0.2 |
GO:2001014 |
regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 |
0.3 |
GO:0016254 |
preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 |
0.0 |
GO:0034182 |
regulation of maintenance of sister chromatid cohesion(GO:0034091) regulation of maintenance of mitotic sister chromatid cohesion(GO:0034182) |
| 0.0 |
0.1 |
GO:0002371 |
dendritic cell cytokine production(GO:0002371) |
| 0.0 |
0.1 |
GO:1903788 |
mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 |
0.1 |
GO:0019075 |
virus maturation(GO:0019075) |
| 0.0 |
0.1 |
GO:0046476 |
glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 |
0.2 |
GO:0015866 |
ADP transport(GO:0015866) |