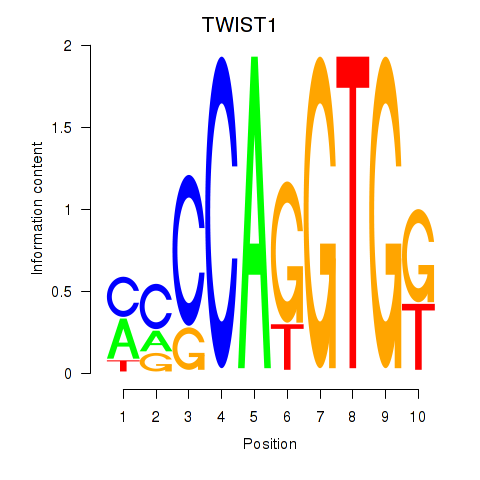
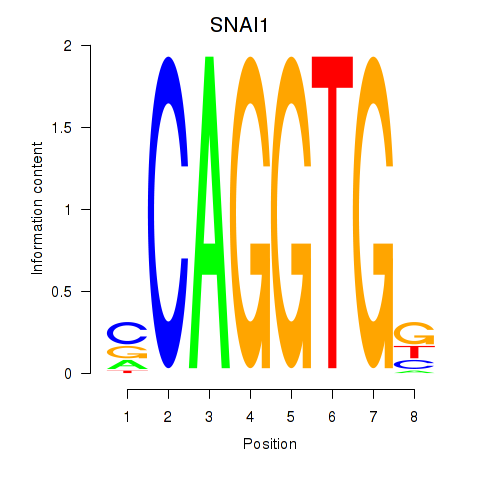
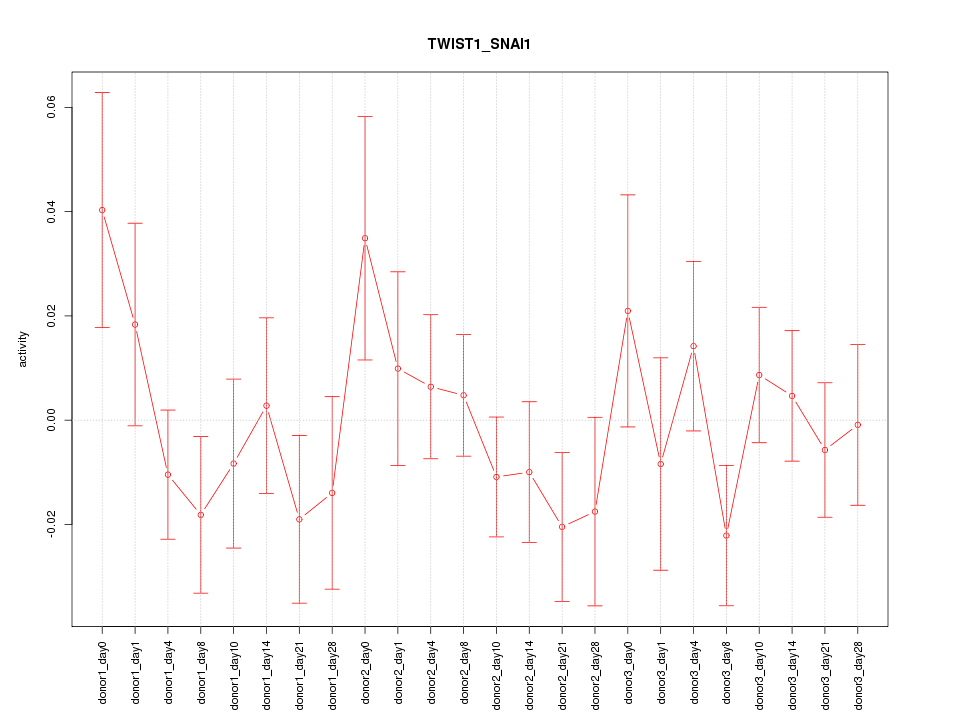
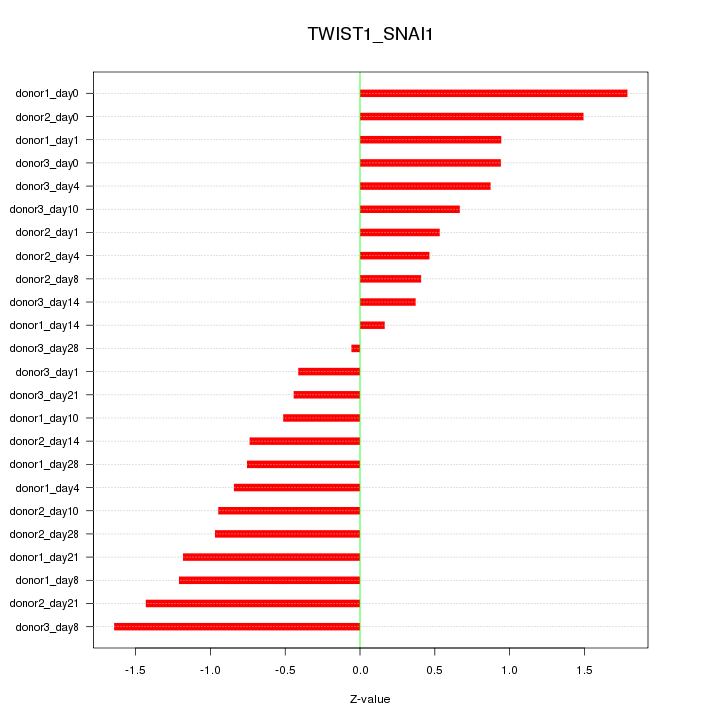
| 0.8 |
2.4 |
GO:0045062 |
extrathymic T cell selection(GO:0045062) |
| 0.6 |
1.8 |
GO:0051780 |
mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.6 |
1.8 |
GO:0031660 |
regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.6 |
1.7 |
GO:1902299 |
pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.6 |
4.5 |
GO:0003065 |
positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.5 |
2.6 |
GO:2000354 |
regulation of ovarian follicle development(GO:2000354) |
| 0.5 |
2.0 |
GO:0010982 |
regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.5 |
2.9 |
GO:0002803 |
positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.4 |
1.3 |
GO:0044209 |
AMP salvage(GO:0044209) |
| 0.4 |
1.2 |
GO:1900085 |
negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.4 |
1.2 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 |
2.0 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.3 |
1.7 |
GO:0003342 |
proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 0.3 |
1.5 |
GO:0009439 |
cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.3 |
6.5 |
GO:0016540 |
protein autoprocessing(GO:0016540) |
| 0.3 |
0.8 |
GO:0032824 |
natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.3 |
1.4 |
GO:1901662 |
phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.3 |
0.8 |
GO:1902910 |
regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.2 |
1.0 |
GO:0003409 |
optic cup structural organization(GO:0003409) |
| 0.2 |
2.8 |
GO:0033227 |
dsRNA transport(GO:0033227) |
| 0.2 |
0.9 |
GO:0032484 |
Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.2 |
2.9 |
GO:0048711 |
positive regulation of astrocyte differentiation(GO:0048711) |
| 0.2 |
1.1 |
GO:0014835 |
myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.2 |
0.7 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) |
| 0.2 |
0.4 |
GO:0021623 |
oculomotor nerve development(GO:0021557) trochlear nerve development(GO:0021558) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.2 |
0.9 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.2 |
0.6 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.2 |
0.2 |
GO:0043000 |
Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.2 |
0.8 |
GO:0007402 |
ganglion mother cell fate determination(GO:0007402) |
| 0.2 |
1.3 |
GO:0051122 |
hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 |
0.5 |
GO:0006500 |
N-terminal protein palmitoylation(GO:0006500) |
| 0.2 |
2.1 |
GO:0061087 |
positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 |
0.3 |
GO:1904862 |
inhibitory synapse assembly(GO:1904862) |
| 0.2 |
0.9 |
GO:0060528 |
secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.2 |
0.5 |
GO:1905166 |
negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.2 |
1.0 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.2 |
0.5 |
GO:1903028 |
positive regulation of opsonization(GO:1903028) |
| 0.2 |
0.7 |
GO:0003164 |
His-Purkinje system development(GO:0003164) |
| 0.2 |
1.8 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.2 |
0.7 |
GO:0010752 |
regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.2 |
1.8 |
GO:0097396 |
response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.2 |
1.6 |
GO:0048733 |
sebaceous gland development(GO:0048733) |
| 0.2 |
0.9 |
GO:0032185 |
septin cytoskeleton organization(GO:0032185) |
| 0.2 |
5.8 |
GO:0034755 |
iron ion transmembrane transport(GO:0034755) |
| 0.2 |
1.1 |
GO:0006642 |
triglyceride mobilization(GO:0006642) |
| 0.1 |
0.4 |
GO:0003169 |
coronary vein morphogenesis(GO:0003169) regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 |
0.4 |
GO:0007219 |
Notch signaling pathway(GO:0007219) |
| 0.1 |
4.0 |
GO:0001829 |
trophectodermal cell differentiation(GO:0001829) |
| 0.1 |
1.3 |
GO:0010820 |
positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 |
1.1 |
GO:0046836 |
glycolipid transport(GO:0046836) |
| 0.1 |
0.8 |
GO:0016479 |
negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 |
0.7 |
GO:1905098 |
negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 |
1.2 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 |
0.5 |
GO:0032811 |
negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.1 |
0.7 |
GO:0021938 |
ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.1 |
0.6 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 |
0.1 |
GO:0039017 |
pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) kidney field specification(GO:0072004) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) |
| 0.1 |
0.8 |
GO:0006982 |
response to lipid hydroperoxide(GO:0006982) |
| 0.1 |
1.4 |
GO:0060075 |
regulation of resting membrane potential(GO:0060075) |
| 0.1 |
1.0 |
GO:1903764 |
regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 |
0.4 |
GO:0010360 |
negative regulation of anion channel activity(GO:0010360) negative regulation of chloride transport(GO:2001226) |
| 0.1 |
0.1 |
GO:0045014 |
carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 |
0.2 |
GO:1903519 |
apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.1 |
0.2 |
GO:0021937 |
cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 |
0.7 |
GO:0061484 |
hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 |
0.3 |
GO:0050894 |
determination of affect(GO:0050894) |
| 0.1 |
0.3 |
GO:0006478 |
peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 |
0.8 |
GO:0021842 |
directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 |
0.3 |
GO:1902630 |
regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 |
0.3 |
GO:0070563 |
negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 0.1 |
0.5 |
GO:0009730 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 |
0.7 |
GO:1902766 |
skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 |
0.2 |
GO:0019087 |
transformation of host cell by virus(GO:0019087) |
| 0.1 |
0.3 |
GO:0035879 |
plasma membrane lactate transport(GO:0035879) |
| 0.1 |
0.5 |
GO:1901503 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 |
1.4 |
GO:0015816 |
glycine transport(GO:0015816) |
| 0.1 |
0.4 |
GO:0046391 |
5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 |
0.5 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 |
0.3 |
GO:0006425 |
glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.1 |
0.3 |
GO:0042369 |
vitamin D catabolic process(GO:0042369) |
| 0.1 |
0.6 |
GO:0071321 |
cellular response to cGMP(GO:0071321) |
| 0.1 |
0.2 |
GO:0002541 |
activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 |
0.3 |
GO:0035359 |
negative regulation of triglyceride catabolic process(GO:0010897) negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 |
0.8 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 0.1 |
0.6 |
GO:1903575 |
cornified envelope assembly(GO:1903575) |
| 0.1 |
1.3 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 |
0.6 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.1 |
0.6 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 |
0.5 |
GO:0033274 |
response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 |
0.5 |
GO:0090402 |
oncogene-induced cell senescence(GO:0090402) |
| 0.1 |
0.3 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 |
0.8 |
GO:0048050 |
post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 |
0.3 |
GO:0048850 |
hypophysis morphogenesis(GO:0048850) |
| 0.1 |
0.2 |
GO:2000646 |
positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 |
0.6 |
GO:0090136 |
epithelial cell-cell adhesion(GO:0090136) |
| 0.1 |
0.3 |
GO:0030264 |
nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.1 |
0.3 |
GO:1990414 |
replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 |
0.3 |
GO:0007343 |
egg activation(GO:0007343) |
| 0.1 |
0.5 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 |
1.3 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 |
1.5 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.1 |
0.5 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) positive regulation of thyroid hormone generation(GO:2000611) |
| 0.1 |
0.4 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 |
0.2 |
GO:0006427 |
histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 |
0.3 |
GO:0043006 |
activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 |
0.3 |
GO:1903936 |
response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.1 |
0.3 |
GO:0060492 |
foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.1 |
0.4 |
GO:1903862 |
positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 |
0.2 |
GO:0071812 |
circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.1 |
0.4 |
GO:0002159 |
desmosome assembly(GO:0002159) |
| 0.1 |
0.5 |
GO:0070384 |
Harderian gland development(GO:0070384) |
| 0.1 |
0.3 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.1 |
0.4 |
GO:0099590 |
neurotransmitter receptor internalization(GO:0099590) |
| 0.1 |
0.2 |
GO:0061073 |
ciliary body morphogenesis(GO:0061073) |
| 0.1 |
0.2 |
GO:0031064 |
negative regulation of histone deacetylation(GO:0031064) |
| 0.1 |
0.3 |
GO:0050712 |
negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 |
0.8 |
GO:2001013 |
epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 |
1.3 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 |
0.4 |
GO:0050428 |
purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 |
0.3 |
GO:0021592 |
fourth ventricle development(GO:0021592) |
| 0.1 |
0.3 |
GO:0001188 |
RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.1 |
0.4 |
GO:0097021 |
Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 |
0.3 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.1 |
3.3 |
GO:0050919 |
negative chemotaxis(GO:0050919) |
| 0.1 |
1.2 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.1 |
0.5 |
GO:2000782 |
regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.1 |
0.4 |
GO:0010193 |
response to ozone(GO:0010193) |
| 0.1 |
1.3 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.1 |
0.4 |
GO:2000667 |
positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.1 |
0.3 |
GO:0055014 |
atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 |
0.3 |
GO:0009726 |
detection of endogenous stimulus(GO:0009726) |
| 0.1 |
0.6 |
GO:0035583 |
sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 |
0.5 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 |
0.1 |
GO:0016539 |
intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 |
0.3 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.1 |
0.1 |
GO:1900106 |
hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 |
0.3 |
GO:0032252 |
secretory granule localization(GO:0032252) |
| 0.1 |
1.0 |
GO:0014041 |
regulation of neuron maturation(GO:0014041) |
| 0.1 |
0.1 |
GO:0000294 |
nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.1 |
0.7 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 |
0.5 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 |
0.2 |
GO:0071484 |
response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.1 |
0.4 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 |
0.3 |
GO:0086073 |
bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 |
0.3 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 |
0.3 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.1 |
0.6 |
GO:2000544 |
cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.1 |
0.3 |
GO:0001306 |
age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 |
0.6 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 |
0.4 |
GO:0007197 |
regulation of vascular smooth muscle contraction(GO:0003056) adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 |
0.2 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.1 |
0.2 |
GO:0033091 |
positive regulation of immature T cell proliferation(GO:0033091) |
| 0.1 |
0.4 |
GO:2001137 |
positive regulation of endocytic recycling(GO:2001137) |
| 0.1 |
0.2 |
GO:0017198 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 |
0.1 |
GO:0051970 |
negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 |
0.2 |
GO:0032911 |
negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 |
7.2 |
GO:0070268 |
cornification(GO:0070268) |
| 0.1 |
0.2 |
GO:0032092 |
positive regulation of protein binding(GO:0032092) |
| 0.1 |
0.5 |
GO:0034201 |
response to oleic acid(GO:0034201) |
| 0.1 |
0.2 |
GO:0009786 |
regulation of asymmetric cell division(GO:0009786) |
| 0.1 |
0.5 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 0.1 |
0.2 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.1 |
0.3 |
GO:0038169 |
somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 |
0.4 |
GO:0055129 |
L-proline biosynthetic process(GO:0055129) |
| 0.1 |
0.3 |
GO:2000382 |
positive regulation of mesoderm development(GO:2000382) |
| 0.1 |
0.2 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.1 |
2.9 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.1 |
0.1 |
GO:1903526 |
negative regulation of membrane tubulation(GO:1903526) |
| 0.1 |
0.2 |
GO:0018352 |
protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 |
0.2 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 |
0.5 |
GO:0030043 |
actin filament fragmentation(GO:0030043) |
| 0.1 |
0.7 |
GO:0035878 |
nail development(GO:0035878) |
| 0.1 |
0.2 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 |
0.6 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.1 |
0.2 |
GO:0010816 |
substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 |
0.2 |
GO:0098912 |
membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.1 |
0.3 |
GO:0043932 |
ossification involved in bone remodeling(GO:0043932) |
| 0.1 |
0.9 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 |
0.1 |
GO:0010544 |
regulation of platelet activation(GO:0010543) negative regulation of platelet activation(GO:0010544) regulation of platelet aggregation(GO:0090330) negative regulation of platelet aggregation(GO:0090331) |
| 0.0 |
1.0 |
GO:0006704 |
glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 |
0.2 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 |
0.8 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.1 |
GO:0006059 |
hexitol metabolic process(GO:0006059) |
| 0.0 |
0.1 |
GO:0048842 |
positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 |
0.1 |
GO:0016999 |
antibiotic metabolic process(GO:0016999) |
| 0.0 |
0.1 |
GO:0032849 |
regulation of cellular pH reduction(GO:0032847) positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 |
1.3 |
GO:0060445 |
branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 |
0.6 |
GO:1903546 |
protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 |
0.3 |
GO:0048702 |
embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 |
0.2 |
GO:0003366 |
cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 |
0.1 |
GO:0071680 |
response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 |
0.1 |
GO:0044376 |
RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 |
0.1 |
GO:0002439 |
chronic inflammatory response to antigenic stimulus(GO:0002439) regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 |
1.0 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 |
0.6 |
GO:0045741 |
positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 |
0.2 |
GO:0030200 |
heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 |
0.5 |
GO:0098887 |
neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 |
0.4 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 |
0.1 |
GO:0003199 |
endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 |
0.4 |
GO:1904352 |
positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 |
0.2 |
GO:0019470 |
4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 |
0.4 |
GO:0021999 |
neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 |
1.0 |
GO:0035767 |
endothelial cell chemotaxis(GO:0035767) |
| 0.0 |
0.2 |
GO:0044501 |
modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 |
0.1 |
GO:1902499 |
positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 |
0.2 |
GO:0010989 |
negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 |
0.2 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 |
0.1 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 |
0.3 |
GO:0031444 |
slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 |
0.8 |
GO:0034656 |
nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 |
0.2 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.0 |
0.1 |
GO:1902227 |
negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.0 |
0.6 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 |
0.8 |
GO:0097369 |
sodium ion import(GO:0097369) |
| 0.0 |
0.9 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.1 |
GO:0060282 |
positive regulation of oocyte development(GO:0060282) |
| 0.0 |
0.6 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 |
0.1 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 |
0.1 |
GO:0099525 |
presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 |
0.2 |
GO:1901725 |
regulation of histone deacetylase activity(GO:1901725) |
| 0.0 |
0.0 |
GO:0050806 |
positive regulation of synaptic transmission(GO:0050806) |
| 0.0 |
1.0 |
GO:0042753 |
positive regulation of circadian rhythm(GO:0042753) |
| 0.0 |
1.4 |
GO:0051482 |
positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 |
0.9 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 |
0.2 |
GO:0033031 |
positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 |
0.3 |
GO:0019344 |
cysteine biosynthetic process(GO:0019344) |
| 0.0 |
0.2 |
GO:1905245 |
regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 |
0.3 |
GO:0098734 |
macromolecule depalmitoylation(GO:0098734) |
| 0.0 |
0.1 |
GO:0044828 |
negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 |
0.5 |
GO:0014067 |
negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 |
0.4 |
GO:1900029 |
positive regulation of ruffle assembly(GO:1900029) |
| 0.0 |
0.3 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.0 |
0.5 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.0 |
0.2 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.0 |
0.1 |
GO:0036292 |
DNA rewinding(GO:0036292) |
| 0.0 |
0.6 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 |
0.1 |
GO:0042421 |
norepinephrine biosynthetic process(GO:0042421) |
| 0.0 |
1.0 |
GO:0014887 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 |
0.3 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 |
0.2 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.1 |
GO:0031630 |
regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 |
0.3 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.2 |
GO:0006824 |
cobalt ion transport(GO:0006824) |
| 0.0 |
0.6 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.1 |
GO:0098746 |
fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 |
0.4 |
GO:0006089 |
lactate metabolic process(GO:0006089) |
| 0.0 |
2.3 |
GO:0019933 |
cAMP-mediated signaling(GO:0019933) |
| 0.0 |
0.4 |
GO:0044351 |
macropinocytosis(GO:0044351) |
| 0.0 |
0.4 |
GO:2000675 |
negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 |
0.2 |
GO:0070980 |
biphenyl catabolic process(GO:0070980) |
| 0.0 |
0.4 |
GO:0098789 |
pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 |
0.1 |
GO:0043376 |
regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 |
1.1 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 |
0.4 |
GO:0009642 |
response to light intensity(GO:0009642) |
| 0.0 |
0.2 |
GO:0060484 |
lung-associated mesenchyme development(GO:0060484) |
| 0.0 |
0.3 |
GO:0060670 |
branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 |
0.1 |
GO:0061760 |
antifungal innate immune response(GO:0061760) |
| 0.0 |
0.5 |
GO:0090179 |
planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 |
0.1 |
GO:1904204 |
regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 |
0.3 |
GO:0061298 |
retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 |
0.1 |
GO:0015917 |
aminophospholipid transport(GO:0015917) |
| 0.0 |
0.1 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 |
0.2 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.0 |
0.3 |
GO:0034465 |
response to carbon monoxide(GO:0034465) smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 |
0.1 |
GO:1902202 |
regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 |
0.2 |
GO:0036337 |
Fas signaling pathway(GO:0036337) necroptotic signaling pathway(GO:0097527) |
| 0.0 |
0.1 |
GO:0006382 |
adenosine to inosine editing(GO:0006382) |
| 0.0 |
0.3 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.0 |
0.1 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.0 |
0.4 |
GO:0051024 |
positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 |
1.1 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.0 |
0.2 |
GO:0007028 |
cytoplasm organization(GO:0007028) |
| 0.0 |
0.1 |
GO:1902474 |
positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 |
0.1 |
GO:0031125 |
rRNA 3'-end processing(GO:0031125) |
| 0.0 |
1.1 |
GO:2000249 |
regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 |
0.5 |
GO:1903204 |
negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 |
1.4 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.0 |
0.3 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 |
0.1 |
GO:1902723 |
negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 |
1.3 |
GO:0050690 |
regulation of defense response to virus by virus(GO:0050690) |
| 0.0 |
0.2 |
GO:0000414 |
regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 |
0.2 |
GO:0008211 |
glucocorticoid metabolic process(GO:0008211) |
| 0.0 |
0.2 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 |
0.2 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 |
0.1 |
GO:0006424 |
glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 |
0.7 |
GO:0035728 |
response to hepatocyte growth factor(GO:0035728) |
| 0.0 |
0.1 |
GO:0010940 |
positive regulation of necrotic cell death(GO:0010940) |
| 0.0 |
0.1 |
GO:0006414 |
translational elongation(GO:0006414) |
| 0.0 |
0.2 |
GO:0019896 |
axonal transport of mitochondrion(GO:0019896) |
| 0.0 |
0.0 |
GO:0018202 |
peptidyl-histidine modification(GO:0018202) |
| 0.0 |
0.2 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 |
0.6 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.7 |
GO:0006884 |
cell volume homeostasis(GO:0006884) |
| 0.0 |
0.1 |
GO:0061762 |
CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 |
0.3 |
GO:0033008 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 |
1.5 |
GO:0006910 |
phagocytosis, recognition(GO:0006910) |
| 0.0 |
0.2 |
GO:0060746 |
maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 |
0.1 |
GO:0060380 |
regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.0 |
0.2 |
GO:0030174 |
regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 |
0.3 |
GO:0090344 |
negative regulation of cell aging(GO:0090344) |
| 0.0 |
0.5 |
GO:0033198 |
response to ATP(GO:0033198) |
| 0.0 |
0.8 |
GO:0007271 |
synaptic transmission, cholinergic(GO:0007271) |
| 0.0 |
0.1 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 |
0.2 |
GO:0070970 |
interleukin-2 secretion(GO:0070970) |
| 0.0 |
0.0 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.1 |
GO:0000738 |
DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 |
0.1 |
GO:2000110 |
negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 |
0.7 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 |
0.2 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.0 |
0.1 |
GO:0032571 |
response to vitamin K(GO:0032571) |
| 0.0 |
0.2 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.0 |
0.1 |
GO:0035811 |
negative regulation of urine volume(GO:0035811) |
| 0.0 |
0.1 |
GO:0071447 |
cellular response to hydroperoxide(GO:0071447) |
| 0.0 |
0.7 |
GO:0090004 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 |
0.1 |
GO:0050760 |
negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 |
0.1 |
GO:0002024 |
diet induced thermogenesis(GO:0002024) |
| 0.0 |
0.1 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 |
0.2 |
GO:1902916 |
positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 |
0.1 |
GO:0035672 |
oligopeptide transmembrane transport(GO:0035672) |
| 0.0 |
0.1 |
GO:1903817 |
negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 |
0.1 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.0 |
0.7 |
GO:0097503 |
sialylation(GO:0097503) |
| 0.0 |
0.7 |
GO:0001954 |
positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 |
1.3 |
GO:0030049 |
muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 |
0.1 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.0 |
0.3 |
GO:0016264 |
gap junction assembly(GO:0016264) |
| 0.0 |
0.6 |
GO:0046627 |
negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 |
1.4 |
GO:1901222 |
regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 |
0.1 |
GO:0038195 |
urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 |
0.3 |
GO:0007224 |
smoothened signaling pathway(GO:0007224) |
| 0.0 |
0.4 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 |
0.5 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.1 |
GO:0006627 |
protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 |
0.3 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 |
1.0 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.0 |
0.7 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.1 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.5 |
GO:0002250 |
adaptive immune response(GO:0002250) |
| 0.0 |
0.1 |
GO:0010801 |
negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 |
0.2 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
0.2 |
GO:0043653 |
mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 |
0.2 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.0 |
0.5 |
GO:0030261 |
chromosome condensation(GO:0030261) |
| 0.0 |
0.3 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.0 |
0.2 |
GO:0046626 |
regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 |
0.2 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.0 |
0.2 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 |
0.1 |
GO:0071493 |
cellular response to UV-B(GO:0071493) |
| 0.0 |
0.1 |
GO:0034334 |
adherens junction maintenance(GO:0034334) |
| 0.0 |
0.3 |
GO:0006044 |
N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 |
0.3 |
GO:0055078 |
sodium ion homeostasis(GO:0055078) |
| 0.0 |
0.3 |
GO:0040018 |
positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 |
0.3 |
GO:0071243 |
cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 |
0.9 |
GO:0045540 |
regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 |
0.1 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.0 |
0.4 |
GO:0030252 |
growth hormone secretion(GO:0030252) |
| 0.0 |
0.5 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
1.7 |
GO:1903955 |
positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 |
0.2 |
GO:1904778 |
regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 |
0.4 |
GO:0051496 |
positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 |
0.9 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.3 |
GO:0002076 |
osteoblast development(GO:0002076) |
| 0.0 |
0.1 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.0 |
0.1 |
GO:0046034 |
ATP metabolic process(GO:0046034) |
| 0.0 |
1.2 |
GO:0006939 |
smooth muscle contraction(GO:0006939) |
| 0.0 |
0.2 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.0 |
0.1 |
GO:0099612 |
protein localization to axon(GO:0099612) |
| 0.0 |
0.5 |
GO:0043616 |
keratinocyte proliferation(GO:0043616) |
| 0.0 |
0.1 |
GO:0030421 |
defecation(GO:0030421) |
| 0.0 |
0.0 |
GO:0032966 |
negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.0 |
0.2 |
GO:0044062 |
regulation of excretion(GO:0044062) |
| 0.0 |
0.2 |
GO:0010644 |
cell communication by electrical coupling(GO:0010644) |
| 0.0 |
0.1 |
GO:0002290 |
gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) positive regulation of gamma-delta T cell activation(GO:0046645) negative regulation of CD8-positive, alpha-beta T cell activation(GO:2001186) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.0 |
0.2 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) |
| 0.0 |
0.2 |
GO:0060707 |
trophoblast giant cell differentiation(GO:0060707) |
| 0.0 |
0.2 |
GO:0042481 |
regulation of odontogenesis(GO:0042481) |
| 0.0 |
0.2 |
GO:0007635 |
chemosensory behavior(GO:0007635) |
| 0.0 |
0.2 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 |
0.2 |
GO:0035092 |
sperm chromatin condensation(GO:0035092) |
| 0.0 |
0.3 |
GO:0003416 |
endochondral bone growth(GO:0003416) |
| 0.0 |
0.1 |
GO:0070673 |
response to interleukin-18(GO:0070673) |
| 0.0 |
0.1 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.1 |
GO:0060828 |
regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 |
0.3 |
GO:0032801 |
receptor catabolic process(GO:0032801) |
| 0.0 |
0.5 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.0 |
0.1 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 |
0.2 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.3 |
GO:0010831 |
positive regulation of myotube differentiation(GO:0010831) |
| 0.0 |
0.7 |
GO:0030834 |
regulation of actin filament depolymerization(GO:0030834) |
| 0.0 |
0.1 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.4 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 |
0.6 |
GO:0030279 |
negative regulation of ossification(GO:0030279) |
| 0.0 |
0.1 |
GO:1903433 |
regulation of constitutive secretory pathway(GO:1903433) |
| 0.0 |
0.6 |
GO:0038083 |
peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 |
0.0 |
GO:0034224 |
cellular response to zinc ion starvation(GO:0034224) |
| 0.0 |
0.2 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.0 |
0.8 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 |
0.1 |
GO:0002903 |
negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 |
0.1 |
GO:0075525 |
viral translational termination-reinitiation(GO:0075525) |
| 0.0 |
0.1 |
GO:0051571 |
positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 |
0.1 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 |
0.1 |
GO:0071816 |
tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 |
0.1 |
GO:0006183 |
GTP biosynthetic process(GO:0006183) |
| 0.0 |
0.3 |
GO:0043928 |
exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 |
0.1 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 |
0.2 |
GO:0010043 |
response to zinc ion(GO:0010043) |
| 0.0 |
0.4 |
GO:0015695 |
organic cation transport(GO:0015695) |
| 0.0 |
0.1 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.2 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.0 |
0.1 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.1 |
GO:0035635 |
entry of bacterium into host cell(GO:0035635) |
| 0.0 |
0.2 |
GO:0060117 |
auditory receptor cell development(GO:0060117) |