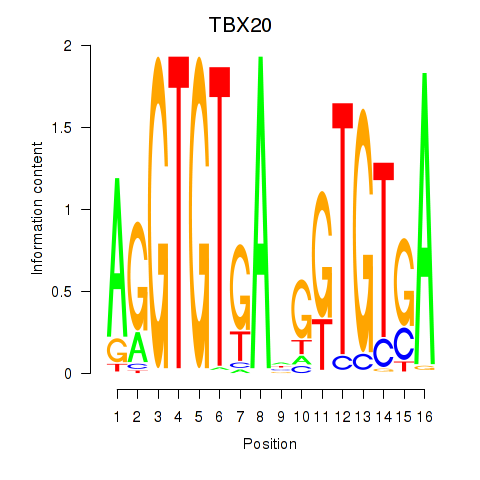
|
chr16_+_57662419
|
0.487
|
ENST00000388812.4
ENST00000538815.1
ENST00000456916.1
ENST00000567154.1
ENST00000388813.5
ENST00000562558.1
ENST00000566271.2
|
GPR56
|
G protein-coupled receptor 56
|
|
chr5_+_6448736
|
0.314
|
ENST00000399816.3
|
UBE2QL1
|
ubiquitin-conjugating enzyme E2Q family-like 1
|
|
chr1_-_11118896
|
0.244
|
ENST00000465788.1
|
SRM
|
spermidine synthase
|
|
chr5_+_135394840
|
0.227
|
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr17_-_7080227
|
0.213
|
ENST00000574330.1
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr1_+_43824669
|
0.197
|
ENST00000372462.1
|
CDC20
|
cell division cycle 20
|
|
chr15_+_76016293
|
0.189
|
ENST00000332145.2
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1
|
|
chr20_+_44509857
|
0.180
|
ENST00000372523.1
ENST00000372520.1
|
ZSWIM1
|
zinc finger, SWIM-type containing 1
|
|
chr12_-_121477039
|
0.166
|
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like
|
|
chr12_-_121476959
|
0.165
|
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like
|
|
chrX_+_67913471
|
0.164
|
ENST00000374597.3
|
STARD8
|
StAR-related lipid transfer (START) domain containing 8
|
|
chr19_+_56915668
|
0.159
|
ENST00000333201.9
ENST00000391778.3
|
ZNF583
|
zinc finger protein 583
|
|
chr7_-_139763521
|
0.149
|
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr6_-_32374900
|
0.147
|
ENST00000374995.3
ENST00000374993.1
ENST00000414363.1
ENST00000540315.1
ENST00000544175.1
ENST00000429232.2
ENST00000454136.3
ENST00000446536.2
|
BTNL2
|
butyrophilin-like 2 (MHC class II associated)
|
|
chr12_+_15475462
|
0.141
|
ENST00000543886.1
ENST00000348962.2
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr6_-_2842087
|
0.112
|
ENST00000537185.1
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr6_-_2842219
|
0.107
|
ENST00000380739.5
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr8_+_27182862
|
0.095
|
ENST00000521164.1
ENST00000346049.5
|
PTK2B
|
protein tyrosine kinase 2 beta
|
|
chr8_+_27183033
|
0.093
|
ENST00000420218.2
|
PTK2B
|
protein tyrosine kinase 2 beta
|
|
chr1_+_11866207
|
0.090
|
ENST00000312413.6
ENST00000346436.6
|
CLCN6
|
chloride channel, voltage-sensitive 6
|
|
chrX_-_52386980
|
0.088
|
ENST00000330906.4
|
XAGE2
|
X antigen family, member 2
|
|
chrX_+_52112158
|
0.085
|
ENST00000286049.2
|
XAGE2B
|
X antigen family, member 2B
|
|
chr12_-_121476750
|
0.083
|
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like
|
|
chr12_-_23737534
|
0.076
|
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr12_+_15475331
|
0.074
|
ENST00000281171.4
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr3_+_186915274
|
0.074
|
ENST00000312295.4
|
RTP1
|
receptor (chemosensory) transporter protein 1
|
|
chr13_+_113760098
|
0.064
|
ENST00000346342.3
ENST00000541084.1
ENST00000375581.3
|
F7
|
coagulation factor VII (serum prothrombin conversion accelerator)
|
|
chr7_-_142207004
|
0.063
|
ENST00000426318.2
|
TRBV10-2
|
T cell receptor beta variable 10-2
|
|
chr20_+_31805131
|
0.061
|
ENST00000375454.3
ENST00000375452.3
|
BPIFA3
|
BPI fold containing family A, member 3
|
|
chr10_-_131762105
|
0.059
|
ENST00000368648.3
ENST00000355311.5
|
EBF3
|
early B-cell factor 3
|
|
chr5_+_161494521
|
0.057
|
ENST00000356592.3
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2
|
|
chr5_+_133861339
|
0.057
|
ENST00000282605.4
ENST00000361895.2
ENST00000402835.1
|
PHF15
|
jade family PHD finger 2
|
|
chr10_+_88414298
|
0.056
|
ENST00000372071.2
|
OPN4
|
opsin 4
|
|
chr2_+_207630081
|
0.056
|
ENST00000236980.6
ENST00000418289.1
ENST00000402774.3
ENST00000403094.3
|
FASTKD2
|
FAST kinase domains 2
|
|
chr10_+_88414338
|
0.054
|
ENST00000241891.5
ENST00000443292.1
|
OPN4
|
opsin 4
|
|
chr1_-_11865982
|
0.051
|
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr11_+_15136462
|
0.048
|
ENST00000379556.3
ENST00000424273.1
|
INSC
|
inscuteable homolog (Drosophila)
|
|
chr10_+_114135952
|
0.044
|
ENST00000356116.1
ENST00000433418.1
ENST00000354273.4
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr3_-_131221790
|
0.043
|
ENST00000512877.1
ENST00000264995.3
ENST00000511168.1
ENST00000425847.2
|
MRPL3
|
mitochondrial ribosomal protein L3
|
|
chr3_+_40428647
|
0.041
|
ENST00000301825.3
ENST00000439533.1
ENST00000456402.1
|
ENTPD3
|
ectonucleoside triphosphate diphosphohydrolase 3
|
|
chr10_+_114135004
|
0.040
|
ENST00000393081.1
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr1_+_162760513
|
0.039
|
ENST00000367915.1
ENST00000367917.3
ENST00000254521.3
ENST00000367913.1
|
HSD17B7
|
hydroxysteroid (17-beta) dehydrogenase 7
|
|
chr17_-_3599492
|
0.038
|
ENST00000435558.1
ENST00000345901.3
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr1_+_22964073
|
0.034
|
ENST00000402322.1
|
C1QA
|
complement component 1, q subcomponent, A chain
|
|
chr17_-_3599327
|
0.034
|
ENST00000551178.1
ENST00000552276.1
ENST00000547178.1
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr19_+_46171464
|
0.029
|
ENST00000590918.1
ENST00000263281.3
ENST00000304207.8
|
GIPR
|
gastric inhibitory polypeptide receptor
|
|
chr17_-_3599696
|
0.028
|
ENST00000225328.5
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr11_-_64703354
|
0.023
|
ENST00000532246.1
ENST00000279168.2
|
GPHA2
|
glycoprotein hormone alpha 2
|
|
chrX_+_7810303
|
0.021
|
ENST00000381059.3
ENST00000341408.4
|
VCX
|
variable charge, X-linked
|
|
chr22_+_38219389
|
0.020
|
ENST00000249041.2
|
GALR3
|
galanin receptor 3
|
|
chrX_+_78200913
|
0.020
|
ENST00000171757.2
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10
|
|
chrX_+_78200829
|
0.019
|
ENST00000544091.1
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10
|
|
chr1_-_17380630
|
0.016
|
ENST00000375499.3
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip)
|
|
chr3_-_50396978
|
0.013
|
ENST00000266025.3
|
TMEM115
|
transmembrane protein 115
|
|
chr1_+_147013182
|
0.011
|
ENST00000234739.3
|
BCL9
|
B-cell CLL/lymphoma 9
|
|
chr2_+_64068074
|
0.010
|
ENST00000394417.2
ENST00000484142.1
ENST00000482668.1
ENST00000467648.2
|
UGP2
|
UDP-glucose pyrophosphorylase 2
|
|
chr7_-_142099977
|
0.006
|
ENST00000390359.3
|
TRBV7-8
|
T cell receptor beta variable 7-8
|
|
chr20_-_48782639
|
0.006
|
ENST00000435301.2
|
RP11-112L6.3
|
RP11-112L6.3
|