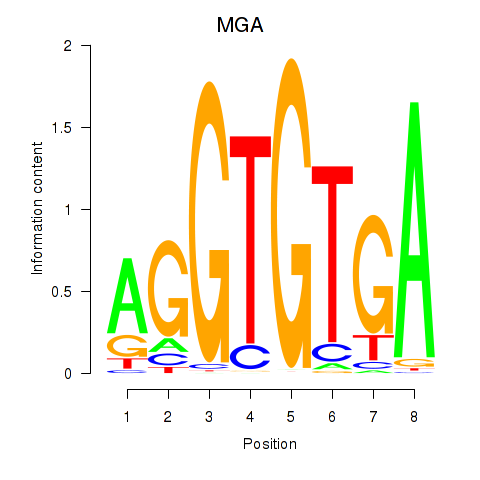
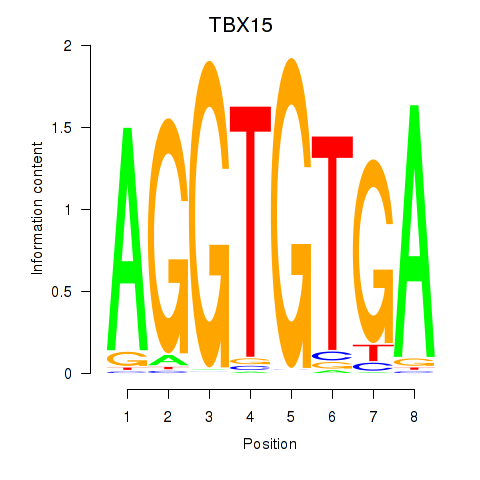
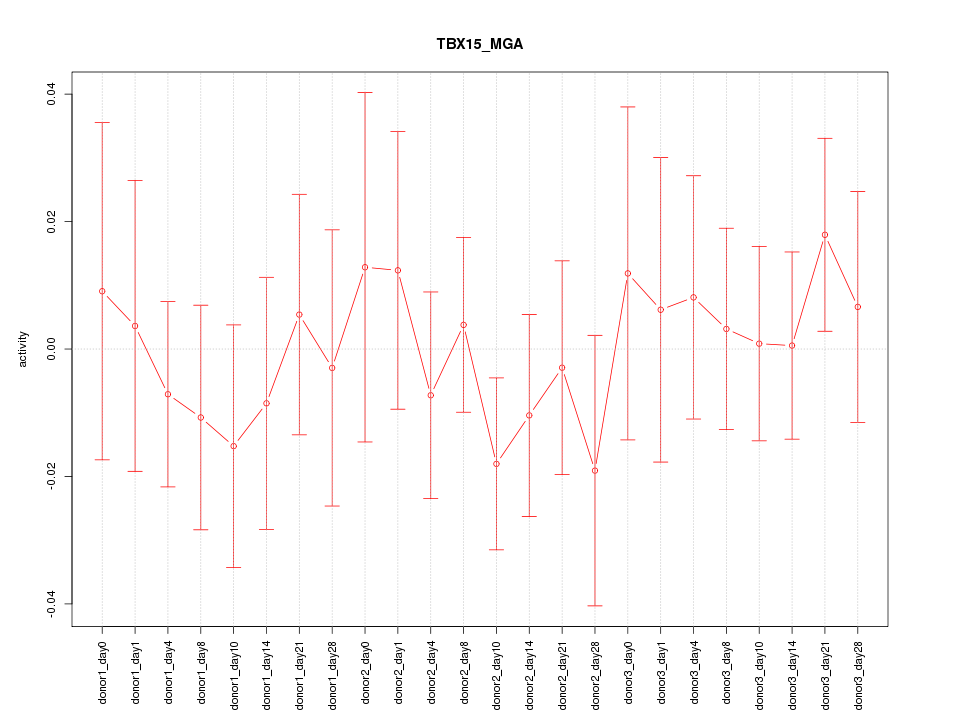
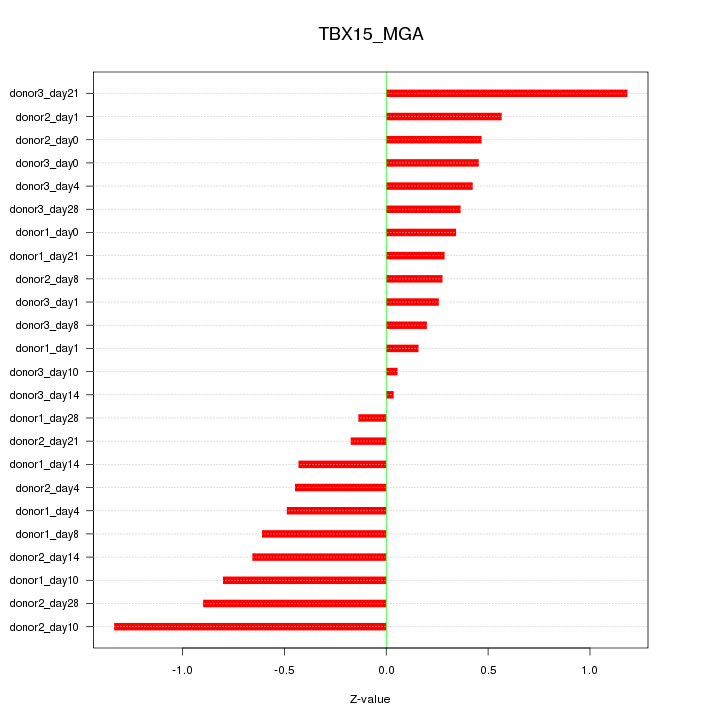
| 0.3 |
0.8 |
GO:0072579 |
molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.3 |
0.8 |
GO:0000412 |
histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.2 |
0.6 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.2 |
0.8 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.2 |
0.8 |
GO:0010982 |
regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.2 |
1.3 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 0.2 |
1.3 |
GO:1903764 |
regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.2 |
0.6 |
GO:0033123 |
positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.2 |
0.9 |
GO:0035803 |
egg coat formation(GO:0035803) |
| 0.2 |
0.5 |
GO:0032824 |
natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 |
0.1 |
GO:2000910 |
negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.1 |
0.4 |
GO:0014839 |
myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.1 |
0.4 |
GO:0097254 |
renal tubular secretion(GO:0097254) |
| 0.1 |
0.5 |
GO:0021633 |
optic nerve structural organization(GO:0021633) |
| 0.1 |
0.3 |
GO:0010652 |
regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 |
0.3 |
GO:0035283 |
rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 |
0.9 |
GO:0038060 |
nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 |
0.3 |
GO:0007412 |
axon target recognition(GO:0007412) |
| 0.1 |
0.3 |
GO:0045014 |
carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 |
0.3 |
GO:1903570 |
regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 |
0.4 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.1 |
0.4 |
GO:0060010 |
Sertoli cell fate commitment(GO:0060010) |
| 0.1 |
1.1 |
GO:0021603 |
cranial nerve formation(GO:0021603) |
| 0.1 |
0.8 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.1 |
0.4 |
GO:0043376 |
regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 |
0.5 |
GO:1901662 |
phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 |
0.3 |
GO:0070563 |
negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 |
0.3 |
GO:0031247 |
actin rod assembly(GO:0031247) |
| 0.1 |
0.3 |
GO:0086053 |
AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.1 |
0.3 |
GO:2000687 |
negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 |
0.4 |
GO:0021938 |
ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.1 |
0.7 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 |
0.2 |
GO:0098838 |
reduced folate transmembrane transport(GO:0098838) |
| 0.1 |
0.5 |
GO:1903575 |
cornified envelope assembly(GO:1903575) |
| 0.1 |
0.9 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
0.4 |
GO:0060574 |
intestinal epithelial cell maturation(GO:0060574) |
| 0.1 |
0.4 |
GO:0043129 |
surfactant homeostasis(GO:0043129) |
| 0.1 |
0.4 |
GO:0019470 |
4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 |
0.3 |
GO:0090526 |
regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 |
0.4 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.1 |
0.2 |
GO:1900195 |
positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 |
0.4 |
GO:0007079 |
mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 |
0.2 |
GO:0050760 |
negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 |
0.2 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.1 |
0.2 |
GO:0002384 |
hepatic immune response(GO:0002384) |
| 0.1 |
0.2 |
GO:0043397 |
corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 |
0.4 |
GO:1902963 |
regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 |
0.3 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 |
0.1 |
GO:0036292 |
DNA rewinding(GO:0036292) |
| 0.1 |
0.3 |
GO:0010728 |
regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.1 |
0.2 |
GO:0044210 |
'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 |
0.2 |
GO:1901876 |
regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 |
0.3 |
GO:0099590 |
neurotransmitter receptor internalization(GO:0099590) |
| 0.1 |
0.2 |
GO:0039019 |
pronephric nephron development(GO:0039019) |
| 0.1 |
0.2 |
GO:0097010 |
eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 |
0.3 |
GO:0010760 |
negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 |
0.2 |
GO:0098759 |
response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.1 |
0.4 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 |
0.2 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 |
0.2 |
GO:2000612 |
pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) thyroid-stimulating hormone secretion(GO:0070460) kidney field specification(GO:0072004) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 |
0.7 |
GO:0036066 |
protein O-linked fucosylation(GO:0036066) |
| 0.1 |
0.3 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 |
0.2 |
GO:0009236 |
cobalamin biosynthetic process(GO:0009236) |
| 0.0 |
1.1 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.0 |
0.3 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.0 |
0.5 |
GO:1903944 |
regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 |
0.3 |
GO:0048859 |
formation of anatomical boundary(GO:0048859) |
| 0.0 |
0.1 |
GO:0007206 |
phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 |
3.2 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.0 |
0.1 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 |
0.1 |
GO:0060474 |
positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 |
0.2 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.0 |
0.2 |
GO:0007343 |
egg activation(GO:0007343) |
| 0.0 |
0.2 |
GO:0008204 |
ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 |
0.1 |
GO:1905205 |
positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 |
0.2 |
GO:0001757 |
somite specification(GO:0001757) |
| 0.0 |
0.2 |
GO:0046035 |
CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 |
0.4 |
GO:0098914 |
membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 |
0.4 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.0 |
0.4 |
GO:0001915 |
negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 |
0.5 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.0 |
0.3 |
GO:0015820 |
leucine transport(GO:0015820) |
| 0.0 |
0.1 |
GO:0055073 |
cadmium ion homeostasis(GO:0055073) divalent metal ion export(GO:0070839) |
| 0.0 |
0.1 |
GO:0070901 |
mitochondrial tRNA methylation(GO:0070901) |
| 0.0 |
0.2 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 |
0.1 |
GO:0014057 |
positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 |
0.3 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.0 |
0.7 |
GO:1990001 |
inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 |
0.2 |
GO:0051461 |
positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 |
0.3 |
GO:0098735 |
positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 |
0.2 |
GO:1904075 |
trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 |
0.5 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) |
| 0.0 |
0.1 |
GO:1903377 |
negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 |
0.3 |
GO:0097646 |
calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 |
0.1 |
GO:0097476 |
spinal cord motor neuron migration(GO:0097476) |
| 0.0 |
0.1 |
GO:1903691 |
positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 |
0.1 |
GO:0003366 |
cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 |
1.2 |
GO:0060216 |
definitive hemopoiesis(GO:0060216) |
| 0.0 |
0.5 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.0 |
GO:0002005 |
angiotensin catabolic process in blood(GO:0002005) |
| 0.0 |
0.2 |
GO:0035879 |
plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.1 |
GO:0072255 |
response to mycophenolic acid(GO:0071505) cellular response to mycophenolic acid(GO:0071506) metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.0 |
0.3 |
GO:0072520 |
seminiferous tubule development(GO:0072520) |
| 0.0 |
0.2 |
GO:0032417 |
positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 |
0.6 |
GO:0001829 |
trophectodermal cell differentiation(GO:0001829) |
| 0.0 |
0.1 |
GO:1990737 |
response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 |
0.1 |
GO:1990637 |
response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.0 |
0.1 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 |
0.1 |
GO:0010966 |
regulation of phosphate transport(GO:0010966) |
| 0.0 |
0.1 |
GO:0070173 |
regulation of enamel mineralization(GO:0070173) |
| 0.0 |
0.5 |
GO:0045954 |
positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 |
0.4 |
GO:0016264 |
gap junction assembly(GO:0016264) |
| 0.0 |
0.2 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 |
0.2 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 |
0.2 |
GO:0070475 |
rRNA base methylation(GO:0070475) |
| 0.0 |
0.4 |
GO:0007213 |
G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 |
1.7 |
GO:0000186 |
activation of MAPKK activity(GO:0000186) |
| 0.0 |
0.1 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.1 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 |
0.1 |
GO:0051582 |
positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 |
0.1 |
GO:0001306 |
age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 |
0.1 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 |
0.1 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.0 |
0.1 |
GO:0001207 |
histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 |
1.0 |
GO:0000289 |
nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 |
0.0 |
GO:2000854 |
positive regulation of corticosterone secretion(GO:2000854) |
| 0.0 |
0.6 |
GO:0034755 |
iron ion transmembrane transport(GO:0034755) |
| 0.0 |
0.1 |
GO:0070676 |
intralumenal vesicle formation(GO:0070676) |
| 0.0 |
0.0 |
GO:0060067 |
cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 |
0.0 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 |
0.2 |
GO:0010666 |
positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 |
0.2 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 |
0.3 |
GO:0010603 |
regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 |
0.1 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.0 |
0.3 |
GO:1904778 |
regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 |
0.1 |
GO:0060215 |
primitive hemopoiesis(GO:0060215) regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 |
0.2 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.0 |
0.2 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.0 |
0.1 |
GO:0043320 |
natural killer cell degranulation(GO:0043320) |
| 0.0 |
0.0 |
GO:0061386 |
closure of optic fissure(GO:0061386) |
| 0.0 |
0.0 |
GO:0010621 |
negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 |
0.2 |
GO:0007585 |
respiratory gaseous exchange(GO:0007585) |
| 0.0 |
0.0 |
GO:0006106 |
fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.0 |
0.0 |
GO:0016185 |
synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 |
0.1 |
GO:0036148 |
phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 |
0.8 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 |
0.2 |
GO:0042487 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 |
0.2 |
GO:0097396 |
response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 |
0.1 |
GO:1901978 |
positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.0 |
0.2 |
GO:0070973 |
protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 |
0.1 |
GO:2000312 |
regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 |
0.3 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 |
0.0 |
GO:0003129 |
heart induction(GO:0003129) |
| 0.0 |
0.0 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.0 |
0.1 |
GO:1903433 |
regulation of constitutive secretory pathway(GO:1903433) |
| 0.0 |
0.2 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.1 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.0 |
0.1 |
GO:2000049 |
positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 |
0.5 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.0 |
0.4 |
GO:0007635 |
chemosensory behavior(GO:0007635) |
| 0.0 |
0.1 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.0 |
0.2 |
GO:0051092 |
positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 |
0.1 |
GO:0097500 |
receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 |
0.1 |
GO:0006824 |
cobalt ion transport(GO:0006824) |
| 0.0 |
0.3 |
GO:0016540 |
protein autoprocessing(GO:0016540) |
| 0.0 |
0.0 |
GO:0051835 |
positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 |
0.3 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 |
0.0 |
GO:0043012 |
regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 |
0.1 |
GO:2000504 |
positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 |
0.0 |
GO:0001994 |
norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 |
0.2 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 |
0.1 |
GO:0051096 |
positive regulation of helicase activity(GO:0051096) |
| 0.0 |
0.4 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
0.1 |
GO:0097475 |
motor neuron migration(GO:0097475) |
| 0.0 |
0.0 |
GO:0014887 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 |
0.1 |
GO:1990564 |
protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 |
0.1 |
GO:0070197 |
meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 |
1.1 |
GO:0050911 |
detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 |
0.2 |
GO:0010592 |
positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 |
0.0 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.0 |
0.1 |
GO:0061113 |
pancreas morphogenesis(GO:0061113) |
| 0.0 |
0.1 |
GO:0000414 |
regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 |
0.2 |
GO:0006954 |
inflammatory response(GO:0006954) |
| 0.0 |
0.1 |
GO:0002667 |
lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 |
0.1 |
GO:0048304 |
positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 |
1.0 |
GO:0007189 |
adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 |
0.4 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.1 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 |
0.1 |
GO:0010793 |
regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 |
0.1 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |