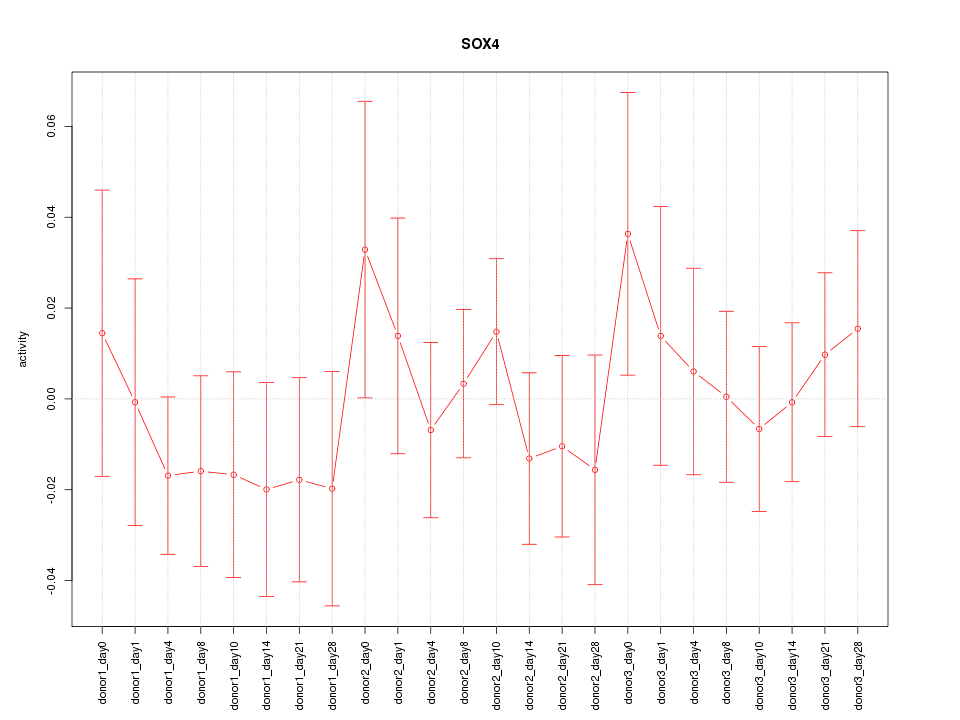
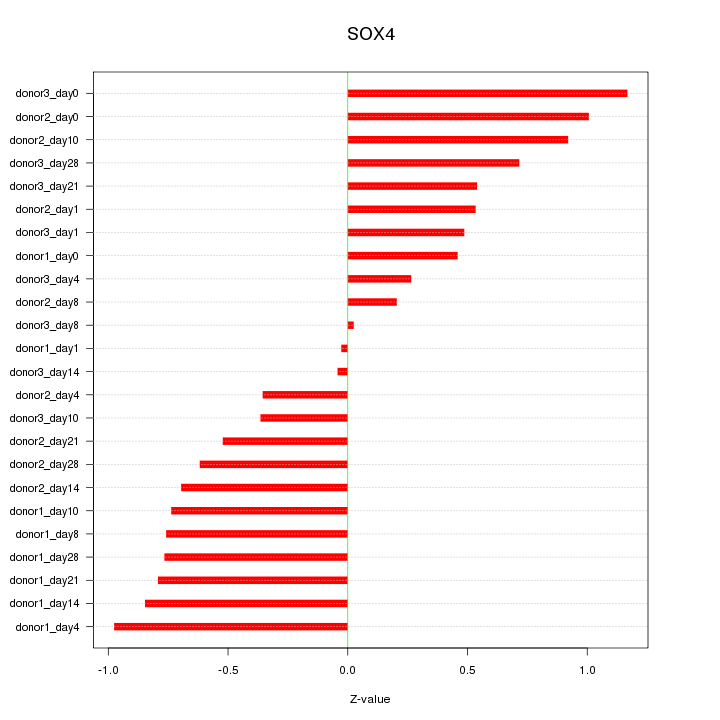
Motif ID: SOX4
Z-value: 0.654
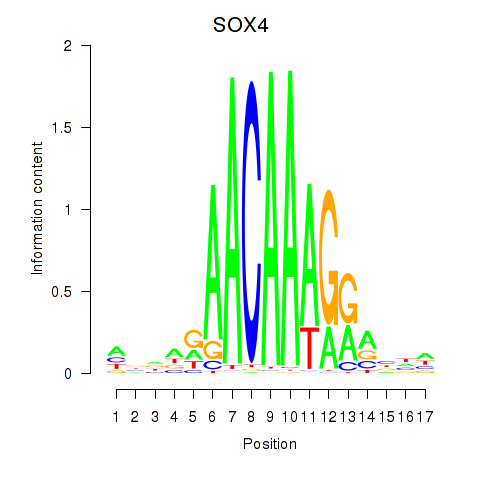
Transcription factors associated with SOX4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| SOX4 | ENSG00000124766.4 | SOX4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX4 | hg19_v2_chr6_+_21593972_21594071 | -0.17 | 4.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.3 | 1.2 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.3 | 1.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.3 | 3.1 | GO:1905031 | regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905031) |
| 0.2 | 0.9 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.2 | 1.6 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.2 | 1.9 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 0.5 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.4 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.4 | GO:0021524 | pulmonary valve formation(GO:0003193) visceral motor neuron differentiation(GO:0021524) foramen ovale closure(GO:0035922) |
| 0.1 | 1.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.8 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.5 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 0.4 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 0.4 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.3 | GO:0048627 | myoblast development(GO:0048627) |
| 0.1 | 0.6 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.5 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.3 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 0.3 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.3 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 0.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 1.0 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 1.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.7 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 1.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.8 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.6 | GO:0035542 | positive regulation of vesicle fusion(GO:0031340) regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.2 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 1.0 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.5 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.6 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.3 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.4 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 1.0 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.7 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.1 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 1.3 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.9 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.5 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.3 | GO:1904262 | TORC2 signaling(GO:0038203) negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 1.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.6 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 1.0 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.3 | 0.9 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.2 | 1.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.2 | 3.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 1.8 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 1.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.9 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.8 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.7 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) bleb(GO:0032059) |
| 0.1 | 0.9 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 1.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.3 | 1.3 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.2 | 1.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.2 | 1.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 0.5 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.2 | 0.5 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.2 | 0.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.8 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.6 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 3.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.4 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.6 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.9 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.1 | 1.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.4 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 1.8 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 2.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 1.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 2.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.3 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.5 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 1.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 0.8 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.9 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.7 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.4 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.3 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.7 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.0 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.2 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.0 | 3.1 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.3 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.8 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.3 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.2 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.8 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.9 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.8 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.5 | REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.4 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.0 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.3 | REACTOME_SIGNALLING_TO_P38_VIA_RIT_AND_RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.4 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.7 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME_DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.3 | REACTOME_POL_SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 1.5 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.1 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.5 | REACTOME_GROWTH_HORMONE_RECEPTOR_SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.6 | REACTOME_MUSCLE_CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.2 | REACTOME_GAP_JUNCTION_TRAFFICKING | Genes involved in Gap junction trafficking |