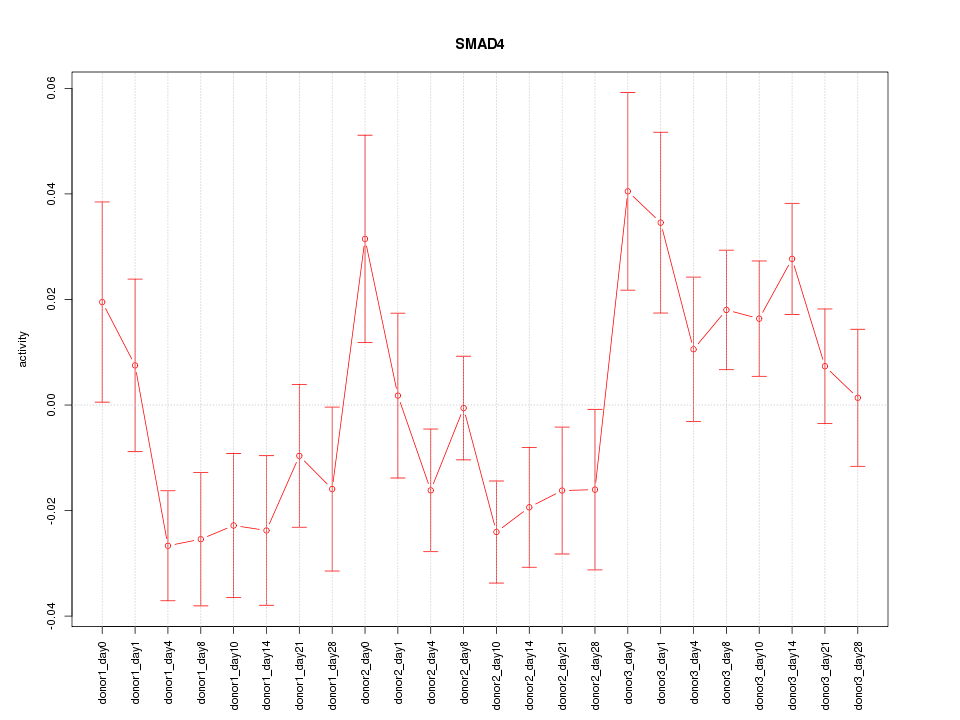
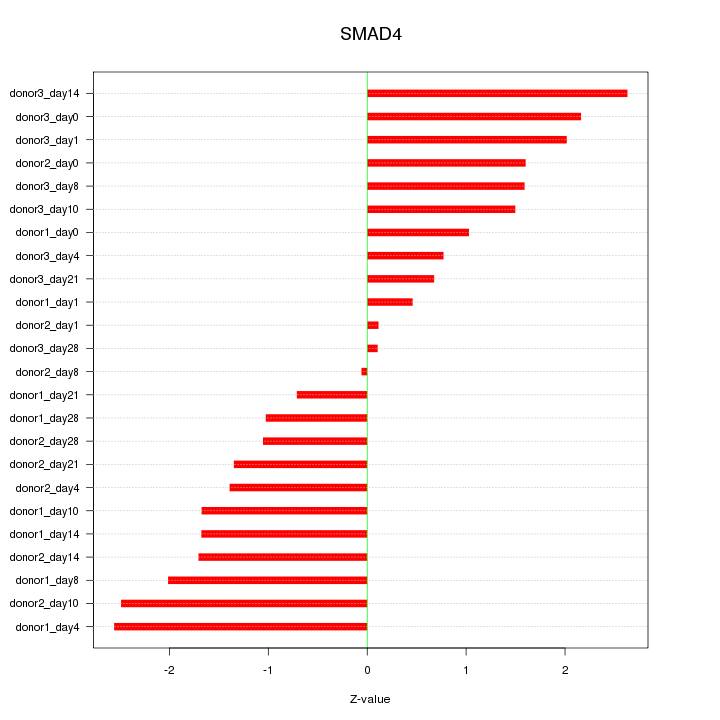
Motif ID: SMAD4
Z-value: 1.543
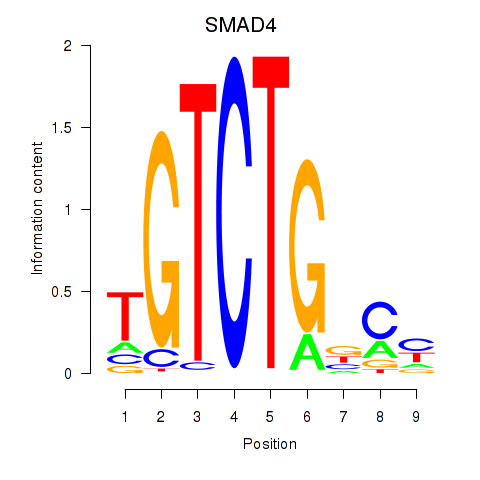
Transcription factors associated with SMAD4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| SMAD4 | ENSG00000141646.9 | SMAD4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMAD4 | hg19_v2_chr18_+_48494361_48494426 | -0.77 | 9.1e-06 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 16.6 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 1.6 | 4.9 | GO:1904298 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 1.2 | 3.6 | GO:0033242 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 1.1 | 4.6 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 1.0 | 3.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 1.0 | 5.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 1.0 | 2.9 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.8 | 2.5 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.8 | 0.8 | GO:0002784 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.7 | 2.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.7 | 2.1 | GO:0002856 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) insulin catabolic process(GO:1901143) |
| 0.6 | 3.8 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.6 | 1.9 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.6 | 1.8 | GO:0052553 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.6 | 1.7 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.5 | 1.6 | GO:1904170 | regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.5 | 1.1 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.5 | 5.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.5 | 1.9 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.5 | 1.4 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.4 | 5.8 | GO:0015816 | glycine transport(GO:0015816) |
| 0.4 | 0.4 | GO:1901205 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) negative regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901205) |
| 0.4 | 1.6 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.4 | 1.2 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.4 | 3.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 1.7 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.3 | 1.0 | GO:0046495 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.3 | 1.3 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.3 | 2.0 | GO:0070458 | cellular detoxification of nitrogen compound(GO:0070458) |
| 0.3 | 2.0 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.3 | 0.3 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.3 | 1.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.3 | 5.6 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.3 | 0.9 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.3 | 1.2 | GO:1902228 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.3 | 1.8 | GO:0042335 | cuticle development(GO:0042335) |
| 0.3 | 1.5 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.3 | 0.9 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.3 | 0.9 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.3 | 1.4 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.3 | 2.2 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.3 | 0.8 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.3 | 0.8 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.3 | 0.8 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.3 | 0.8 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.3 | 2.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.3 | 1.0 | GO:0051884 | anagen(GO:0042640) regulation of anagen(GO:0051884) |
| 0.3 | 1.0 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 | 1.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.3 | 1.5 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.2 | 0.7 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.2 | 1.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.2 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 0.9 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.2 | 1.2 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular response to UV-A(GO:0071492) |
| 0.2 | 1.2 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.2 | 0.9 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 0.7 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.2 | 1.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 0.5 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.2 | 2.0 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.2 | 0.4 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.2 | 1.3 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.2 | 0.6 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.2 | 1.0 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 1.6 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.2 | 2.4 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.2 | 1.6 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.2 | 1.7 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 1.4 | GO:0070305 | response to cGMP(GO:0070305) |
| 0.2 | 0.8 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.2 | 0.6 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 0.2 | 1.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.2 | 1.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 1.3 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.2 | 0.7 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.2 | 0.5 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.2 | 0.4 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.2 | 0.9 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 1.1 | GO:0061364 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) response to cortisol(GO:0051414) apoptotic process involved in luteolysis(GO:0061364) |
| 0.2 | 0.9 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.2 | 0.7 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.2 | 0.5 | GO:0060545 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) positive regulation of necroptotic process(GO:0060545) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.2 | 0.7 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 0.3 | GO:0071672 | negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.2 | 0.5 | GO:0050894 | determination of affect(GO:0050894) |
| 0.2 | 0.8 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.2 | 2.4 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 | 0.8 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.2 | 0.8 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 0.8 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.2 | 0.6 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.2 | 0.6 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 0.6 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.2 | 0.5 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 0.4 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 0.7 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.1 | 2.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 1.0 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.7 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.4 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.4 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.7 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.8 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 0.8 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.4 | GO:2000910 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.1 | 0.8 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.9 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.1 | 0.5 | GO:0072376 | protein activation cascade(GO:0072376) |
| 0.1 | 0.7 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.1 | 1.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 0.8 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.1 | 0.8 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.7 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 | 0.4 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.1 | 0.5 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.6 | GO:0048749 | compound eye development(GO:0048749) |
| 0.1 | 0.4 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.1 | 1.8 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.1 | 1.0 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 1.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.4 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.1 | 1.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 0.4 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 1.2 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 | 0.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 1.6 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 0.5 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.1 | 0.3 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.1 | 1.1 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 1.0 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 0.7 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.1 | 2.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 1.3 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.3 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.1 | 0.8 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 5.0 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.4 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.1 | 0.4 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 4.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 2.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.5 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 1.6 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.8 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.2 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.3 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 1.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.6 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 1.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.2 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 0.3 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.1 | 0.3 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.1 | 0.3 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) response to capsazepine(GO:1901594) |
| 0.1 | 0.8 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.4 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.1 | 0.3 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.9 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.5 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.4 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.4 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 0.4 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.1 | 1.5 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.4 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.3 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.1 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.3 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.3 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.1 | 0.7 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.3 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 1.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.3 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.1 | 0.4 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 | 0.2 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 1.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 0.4 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.2 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 1.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.2 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.1 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 0.3 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.4 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.1 | 0.2 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.1 | 0.3 | GO:0051493 | regulation of cytoskeleton organization(GO:0051493) |
| 0.1 | 0.5 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.1 | 0.2 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.2 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 1.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 3.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.9 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.1 | 0.3 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.5 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 0.4 | GO:0050668 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.1 | 2.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.7 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 0.1 | 9.0 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.3 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.1 | 1.0 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 1.4 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.1 | 0.3 | GO:0007276 | gamete generation(GO:0007276) |
| 0.1 | 0.9 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.7 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 2.2 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.5 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 0.2 | GO:1904808 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.1 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.3 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 0.5 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 7.4 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 0.6 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.1 | 0.2 | GO:2001137 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 0.1 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.1 | 1.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.5 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 1.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 0.3 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.1 | 0.6 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.4 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.6 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.3 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 0.8 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 1.9 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.4 | GO:0046643 | regulation of gamma-delta T cell activation(GO:0046643) |
| 0.1 | 0.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.4 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.2 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.1 | 1.3 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 0.2 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.1 | 0.2 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.1 | 1.6 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 0.5 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.1 | 1.0 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 | 0.5 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 1.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.2 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 0.3 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.2 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.3 | GO:0015824 | proline transport(GO:0015824) proline transmembrane transport(GO:0035524) |
| 0.0 | 1.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.4 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.1 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.3 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.0 | 0.9 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.8 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.7 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0060921 | sinoatrial node development(GO:0003163) sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.3 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.5 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.7 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.0 | 0.5 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 2.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.5 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.0 | 0.3 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.1 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.4 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.5 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.5 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.3 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.3 | GO:0006366 | transcription from RNA polymerase II promoter(GO:0006366) |
| 0.0 | 1.8 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.3 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.4 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.4 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.5 | GO:0006935 | chemotaxis(GO:0006935) taxis(GO:0042330) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.9 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.9 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.2 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.2 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.6 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.4 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.6 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 1.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.4 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 2.3 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.2 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 1.2 | GO:0071174 | mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.1 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 2.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 1.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.5 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.0 | 0.3 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.1 | GO:0051135 | positive regulation of NK T cell activation(GO:0051135) |
| 0.0 | 0.0 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 1.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.0 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.5 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.7 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 1.4 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 1.5 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0051969 | regulation of transmission of nerve impulse(GO:0051969) |
| 0.0 | 0.8 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 | 0.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.8 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.9 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.0 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.4 | GO:0010919 | regulation of inositol phosphate biosynthetic process(GO:0010919) positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.4 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.2 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.9 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.0 | 0.1 | GO:0035814 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 | 0.5 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.5 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.4 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.9 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.5 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.0 | 0.1 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 0.6 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.4 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.4 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.4 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.4 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.3 | GO:0007015 | actin filament organization(GO:0007015) |
| 0.0 | 0.1 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.2 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.0 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.0 | 1.2 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.5 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.2 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.4 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.3 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.0 | GO:0060481 | bronchus development(GO:0060433) lobar bronchus epithelium development(GO:0060481) lobar bronchus development(GO:0060482) |
| 0.0 | 0.2 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 1.4 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 0.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.4 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.8 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.6 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 | 0.4 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0009227 | UDP-N-acetylglucosamine catabolic process(GO:0006049) nucleotide-sugar catabolic process(GO:0009227) |
| 0.0 | 0.6 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 0.1 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.1 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.0 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 0.0 | 0.1 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.0 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.4 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 0.3 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.1 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.6 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.3 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.2 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.2 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 1.2 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 | 0.2 | GO:0072678 | T cell migration(GO:0072678) |
| 0.0 | 0.1 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.5 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.5 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 17.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 1.1 | 3.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.8 | 2.5 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.5 | 1.8 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.4 | 0.8 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.4 | 1.4 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.3 | 1.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.3 | 7.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 0.8 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.3 | 4.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 0.9 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 0.7 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) other organism cytoplasm(GO:0097679) |
| 0.2 | 1.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.2 | 0.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.2 | 1.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 0.7 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.2 | 1.4 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 1.0 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 0.6 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.2 | 0.5 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.1 | 0.9 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.5 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.8 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 3.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.5 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 1.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 1.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 2.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.7 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 1.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.7 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.3 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.1 | 2.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 5.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 3.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.3 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.1 | 0.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.8 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.6 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.8 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 0.4 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.1 | 0.3 | GO:0043231 | intracellular membrane-bounded organelle(GO:0043231) |
| 0.1 | 1.7 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 0.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.4 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 9.4 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 0.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.3 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.7 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.1 | 2.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.8 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 1.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 0.7 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 7.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.5 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.1 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.1 | 0.4 | GO:0034448 | EGO complex(GO:0034448) |
| 0.1 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 1.5 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 3.5 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.6 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:1990742 | microvesicle(GO:1990742) |
| 0.0 | 6.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 2.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 3.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.8 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 2.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 2.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.0 | GO:0070161 | anchoring junction(GO:0070161) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 2.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 8.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 1.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 3.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.0 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 11.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.3 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 1.0 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 1.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 7.5 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.5 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.4 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 1.0 | 5.2 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 1.0 | 4.9 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.8 | 3.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.7 | 3.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.5 | 5.8 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.5 | 5.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.5 | 1.9 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.5 | 1.8 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.4 | 2.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.4 | 2.0 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.4 | 2.4 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.4 | 2.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.4 | 5.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.4 | 1.5 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.4 | 1.1 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.3 | 1.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 1.4 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.3 | 3.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 1.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.3 | 5.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.3 | 2.4 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.3 | 1.5 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.3 | 0.9 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.3 | 0.8 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.3 | 3.5 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.2 | 1.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 1.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.2 | 1.2 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.2 | 1.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 0.7 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.2 | 0.7 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.2 | 2.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 5.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 3.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 1.5 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 0.6 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 1.5 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 0.8 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 0.6 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 0.8 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.2 | 1.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 1.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.2 | 1.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.2 | 0.7 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.2 | 1.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.2 | 0.7 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 2.0 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 1.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.2 | 1.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.2 | 1.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.2 | 1.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 1.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 0.5 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.2 | 0.7 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 0.5 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.2 | 1.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 0.8 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 2.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 1.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.5 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.1 | 1.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.5 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.4 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 0.5 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 0.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.4 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 3.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.4 | GO:0051990 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.1 | 0.8 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.7 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.3 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.1 | 0.3 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.5 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.6 | GO:0042835 | BRE binding(GO:0042835) |
| 0.1 | 0.3 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 0.7 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.4 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.3 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) tetrahydrobiopterin binding(GO:0034617) |
| 0.1 | 0.4 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 2.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 1.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.3 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 0.8 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 0.1 | 1.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 3.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.7 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 0.3 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.2 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.8 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 2.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.4 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.6 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 1.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.9 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.3 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 2.0 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.2 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 2.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.2 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.1 | 1.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 2.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.3 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 0.3 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 1.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.4 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 2.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.5 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.9 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.6 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 0.3 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 0.6 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 1.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 5.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.3 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 0.2 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.1 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 2.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 15.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 1.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 1.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 1.0 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 1.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 1.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.7 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.5 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.6 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 1.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.8 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 1.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.7 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 1.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 2.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 3.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 1.6 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 1.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.6 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 1.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.6 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.4 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 2.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 1.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 1.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.3 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 2.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 2.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 1.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 2.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0017016 | Ras GTPase binding(GO:0017016) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 4.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 2.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.7 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:0031078 | NAD-dependent histone deacetylase activity(GO:0017136) histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 1.3 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.1 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 4.4 | PID_S1P_S1P1_PATHWAY | S1P1 pathway |
| 0.1 | 2.5 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.9 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 0.6 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 6.1 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.1 | 3.8 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 4.3 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.1 | 4.7 | PID_INTEGRIN1_PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 5.3 | PID_TCR_PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 2.7 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.6 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.3 | SA_TRKA_RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.9 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.6 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.4 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.7 | PID_CD8_TCR_DOWNSTREAM_PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 3.7 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 1.2 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 2.3 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.2 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.9 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 1.7 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | PID_IL8_CXCR2_PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.5 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.0 | 1.2 | PID_AMB2_NEUTROPHILS_PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.2 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.9 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.4 | PID_ENDOTHELIN_PATHWAY | Endothelins |
| 0.0 | 0.9 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 2.2 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.1 | PID_S1P_S1P2_PATHWAY | S1P2 pathway |
| 0.0 | 0.5 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.7 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.7 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | ST_DIFFERENTIATION_PATHWAY_IN_PC12_CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.5 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
| 0.0 | 1.3 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID_AR_NONGENOMIC_PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.0 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.6 | PID_VEGFR1_2_PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.0 | 3.8 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID_PI3K_PLC_TRK_PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.5 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.0 | 0.6 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.3 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.0 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 7.9 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 5.3 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.3 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.0 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 4.6 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 4.1 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.1 | 2.2 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 3.2 | REACTOME_REGULATION_OF_IFNA_SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 2.3 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.4 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 3.4 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.9 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 0.5 | REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.7 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.5 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.4 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 1.0 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.2 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.9 | REACTOME_SIGNAL_TRANSDUCTION_BY_L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 0.9 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.9 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 4.8 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 3.7 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.5 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.0 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.8 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.7 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.8 | REACTOME_REVERSIBLE_HYDRATION_OF_CARBON_DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.3 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.6 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 3.5 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.9 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.6 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.7 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.4 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.7 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.9 | REACTOME_SIGNALING_BY_CONSTITUTIVELY_ACTIVE_EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.7 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.9 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.1 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.3 | REACTOME_INHIBITION_OF_THE_PROTEOLYTIC_ACTIVITY_OF_APC_C_REQUIRED_FOR_THE_ONSET_OF_ANAPHASE_BY_MITOTIC_SPINDLE_CHECKPOINT_COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.6 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.8 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME_ACTIVATION_OF_NF_KAPPAB_IN_B_CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.6 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.0 | 2.1 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.5 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME_FORMATION_OF_TUBULIN_FOLDING_INTERMEDIATES_BY_CCT_TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.7 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME_APOBEC3G_MEDIATED_RESISTANCE_TO_HIV1_INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.6 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 2.2 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.4 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.0 | 1.1 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.8 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.8 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME_TRYPTOPHAN_CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 2.8 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.1 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.4 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME_INHIBITION_OF_INSULIN_SECRETION_BY_ADRENALINE_NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.1 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME_INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.6 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.1 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.0 | REACTOME_CELL_SURFACE_INTERACTIONS_AT_THE_VASCULAR_WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.1 | REACTOME_SOS_MEDIATED_SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.5 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.4 | REACTOME_RIP_MEDIATED_NFKB_ACTIVATION_VIA_DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 1.4 | REACTOME_CROSS_PRESENTATION_OF_SOLUBLE_EXOGENOUS_ANTIGENS_ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.1 | REACTOME_NITRIC_OXIDE_STIMULATES_GUANYLATE_CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.0 | 1.6 | REACTOME_TRANS_GOLGI_NETWORK_VESICLE_BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.3 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME_G_PROTEIN_ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 3.5 | REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.2 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.6 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME_IL_2_SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.2 | REACTOME_TRAFFICKING_AND_PROCESSING_OF_ENDOSOMAL_TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.6 | REACTOME_PHASE_II_CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.0 | REACTOME_CELL_JUNCTION_ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.3 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.1 | REACTOME_ADENYLATE_CYCLASE_INHIBITORY_PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_GLUCAGON_LIKE_PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.4 | REACTOME_STEROID_HORMONES | Genes involved in Steroid hormones |