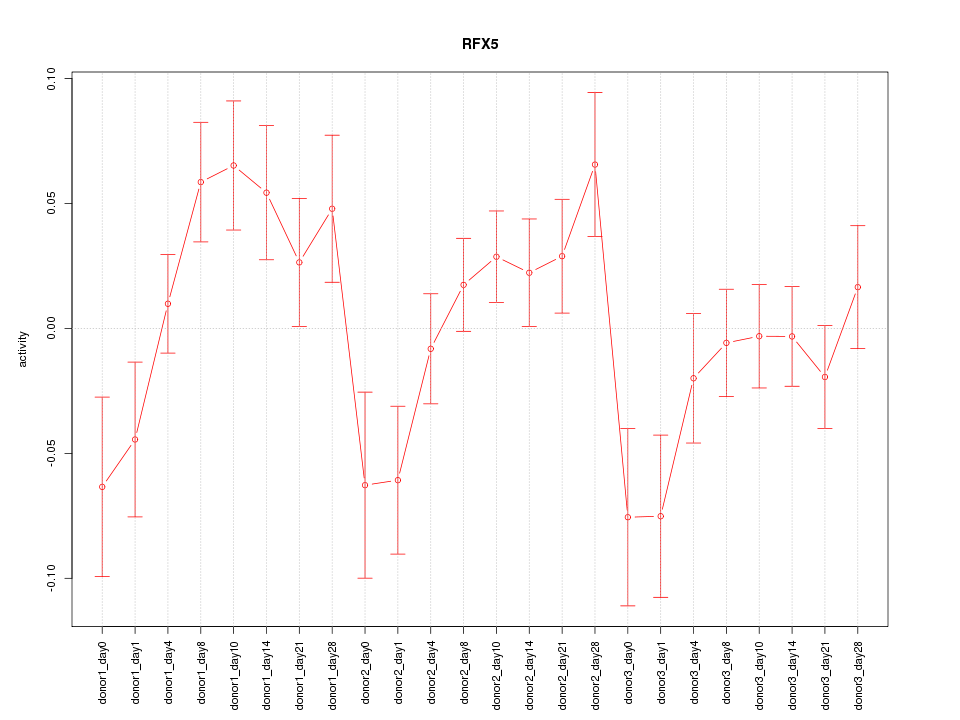
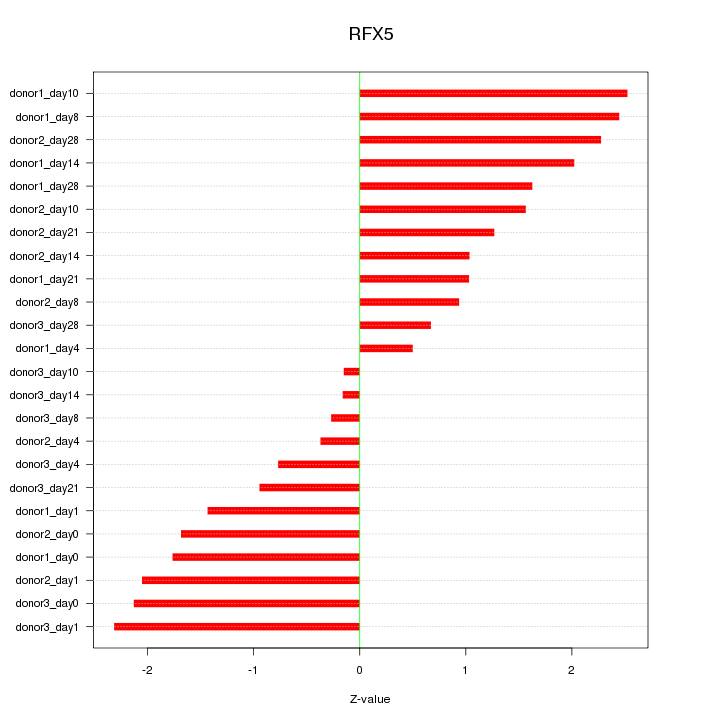
Motif ID: RFX5
Z-value: 1.526
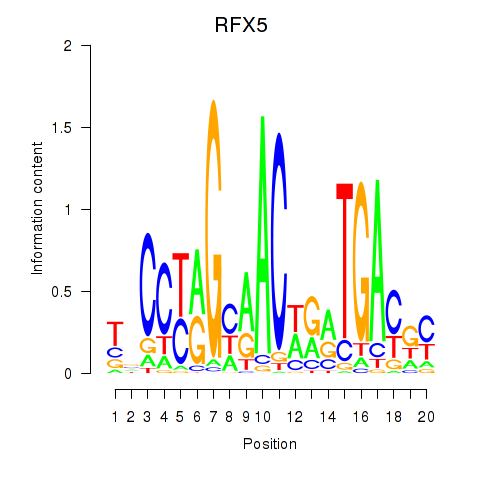
Transcription factors associated with RFX5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| RFX5 | ENSG00000143390.13 | RFX5 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX5 | hg19_v2_chr1_-_151319710_151319833 | 0.45 | 2.9e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 29.6 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 2.7 | 13.5 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 1.1 | 14.9 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.8 | 2.3 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.3 | 1.0 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.3 | 4.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 31.5 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.2 | 4.3 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.2 | 0.8 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.2 | 0.5 | GO:1903989 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.2 | 7.5 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.1 | 1.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 14.0 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.1 | 10.8 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 4.0 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 0.4 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 1.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.6 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.1 | 3.9 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 1.0 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 2.7 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 1.2 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 0.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.0 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.3 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.7 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 1.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 5.5 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.2 | GO:0048733 | peripheral nervous system myelin maintenance(GO:0032287) sebaceous gland development(GO:0048733) |
| 0.0 | 1.7 | GO:0002456 | T cell mediated immunity(GO:0002456) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 73.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.6 | 14.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.7 | 14.9 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.6 | 10.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 3.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.5 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 5.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 4.1 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 4.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 2.5 | GO:1902911 | protein kinase complex(GO:1902911) |
| 0.0 | 2.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 13.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.2 | 10.8 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 1.1 | 42.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.8 | 4.0 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.6 | 14.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.5 | 13.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.5 | 14.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.4 | 1.1 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.3 | 5.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 1.3 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.2 | 1.0 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 2.3 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 1.0 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 4.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.3 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.8 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 1.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 3.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 16.2 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.3 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.1 | PID_CD40_PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 37.9 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 14.0 | REACTOME_CROSS_PRESENTATION_OF_SOLUBLE_EXOGENOUS_ANTIGENS_ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.2 | 25.6 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 0.5 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.5 | REACTOME_AUTODEGRADATION_OF_CDH1_BY_CDH1_APC_C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 2.3 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.3 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |