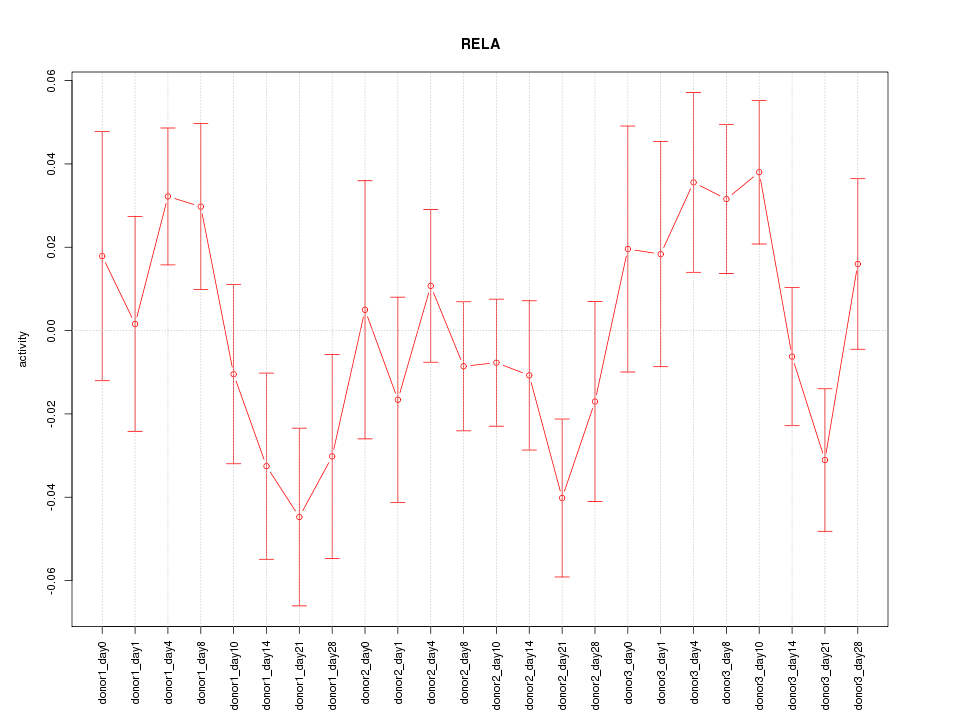
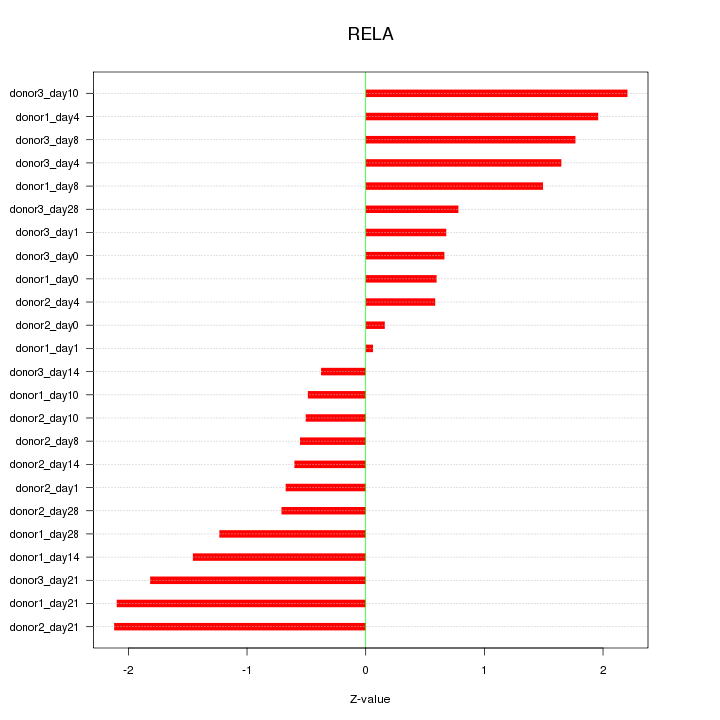
Motif ID: RELA
Z-value: 1.244
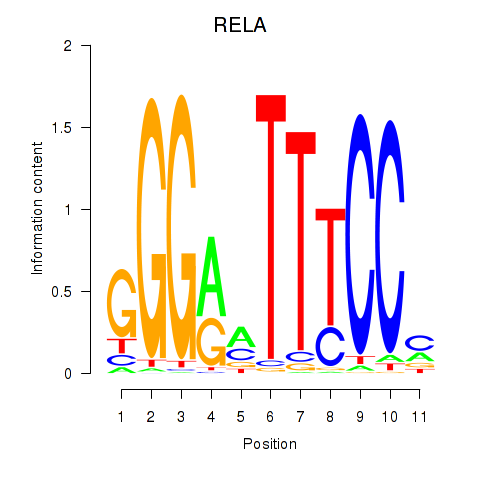
Transcription factors associated with RELA:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| RELA | ENSG00000173039.14 | RELA |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RELA | hg19_v2_chr11_-_65430251_65430399 | 0.55 | 5.5e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 1.2 | 3.6 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.6 | 2.5 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.6 | 1.7 | GO:0070428 | B-1 B cell homeostasis(GO:0001922) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.6 | 1.7 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.5 | 1.6 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.5 | 4.4 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.5 | 1.4 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.4 | 1.8 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.4 | 1.3 | GO:0071790 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.4 | 8.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.3 | 1.0 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.3 | 1.4 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.3 | 2.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.3 | 1.6 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.3 | 3.0 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.3 | 1.0 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.2 | 1.1 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.2 | 0.6 | GO:2000364 | hepatic immune response(GO:0002384) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.2 | 1.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 0.6 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.2 | 1.8 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.2 | 0.6 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.2 | 0.6 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.2 | 0.6 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.2 | 1.3 | GO:2000669 | positive regulation of pinocytosis(GO:0048549) negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.2 | 0.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 1.6 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.2 | 0.9 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.2 | 0.8 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.2 | 1.4 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 0.6 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.1 | 0.6 | GO:1902228 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.1 | 0.6 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 1.0 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 3.0 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 2.1 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.7 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.3 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.1 | 0.3 | GO:0031938 | regulation of chromatin silencing at telomere(GO:0031938) |
| 0.1 | 0.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 1.1 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 2.0 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.4 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 1.5 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.4 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 1.0 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.7 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 3.3 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 1.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.3 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.5 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.8 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 1.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 2.4 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.3 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 1.1 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.1 | 1.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.2 | GO:1903347 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 1.9 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.4 | GO:0036343 | psychomotor behavior(GO:0036343) motor behavior(GO:0061744) |
| 0.1 | 0.2 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
| 0.1 | 0.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.5 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 1.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 1.4 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.0 | 0.6 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.5 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.3 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.3 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.3 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 1.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.5 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 1.1 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 1.6 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.1 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.5 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.4 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:2000551 | regulation of T-helper 2 cell cytokine production(GO:2000551) |
| 0.0 | 1.7 | GO:0060113 | inner ear receptor cell differentiation(GO:0060113) |
| 0.0 | 1.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.8 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:1904529 | regulation of actin filament binding(GO:1904529) negative regulation of actin filament binding(GO:1904530) regulation of actin binding(GO:1904616) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 2.8 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.1 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.5 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.7 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.4 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.4 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.4 | 1.3 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.4 | 1.6 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.3 | 0.8 | GO:0071749 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.2 | 1.8 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 1.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 2.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 2.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.6 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.3 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 1.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.3 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.2 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.1 | 1.3 | GO:0033643 | host cell part(GO:0033643) |
| 0.1 | 0.9 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 1.4 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.9 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 2.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 3.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 4.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 7.8 | GO:0045121 | membrane raft(GO:0045121) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0071204 | U7 snRNP(GO:0005683) histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 21.0 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.9 | 3.6 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.7 | 11.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.5 | 2.4 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.2 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.2 | 1.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 1.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 0.6 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 1.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 0.6 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 0.4 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.1 | 2.0 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 1.6 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 1.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.8 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.3 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 2.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 1.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 2.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 2.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.3 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 1.0 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.2 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.1 | 0.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.6 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 2.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 1.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 1.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 1.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.3 | GO:0010851 | cyclase regulator activity(GO:0010851) |
| 0.0 | 0.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 3.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 8.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 1.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 2.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 2.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 4.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.9 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 8.5 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.1 | 3.6 | PID_S1P_S1P1_PATHWAY | S1P1 pathway |
| 0.1 | 3.2 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 2.9 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.8 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.2 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.4 | PID_ECADHERIN_KERATINOCYTE_PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.2 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.7 | PID_CXCR3_PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.5 | PID_NFKAPPAB_ATYPICAL_PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 2.0 | PID_AMB2_NEUTROPHILS_PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.5 | ST_TYPE_I_INTERFERON_PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.1 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.6 | PID_IL8_CXCR2_PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.5 | ST_INTERLEUKIN_4_PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 2.9 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.2 | PID_TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 11.4 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.2 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.7 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.0 | 0.8 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.2 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 2.0 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | PID_ENDOTHELIN_PATHWAY | Endothelins |
| 0.0 | 0.8 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.8 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID_BCR_5PATHWAY | BCR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 16.0 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 7.5 | REACTOME_RIP_MEDIATED_NFKB_ACTIVATION_VIA_DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 2.7 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 5.1 | REACTOME_NOD1_2_SIGNALING_PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.0 | REACTOME_TRAF6_MEDIATED_NFKB_ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 2.3 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.1 | REACTOME_TRAF3_DEPENDENT_IRF_ACTIVATION_PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.9 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 3.6 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.3 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.5 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.0 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.5 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 2.2 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 3.6 | REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.0 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.6 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.9 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME_NFKB_ACTIVATION_THROUGH_FADD_RIP1_PATHWAY_MEDIATED_BY_CASPASE_8_AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.3 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.6 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 2.1 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME_CD28_DEPENDENT_PI3K_AKT_SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.8 | REACTOME_MUSCLE_CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 1.5 | REACTOME_CELL_JUNCTION_ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.9 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.6 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |