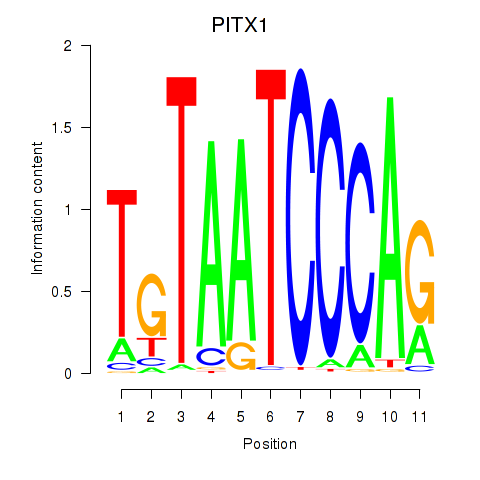
Motif ID: PITX1
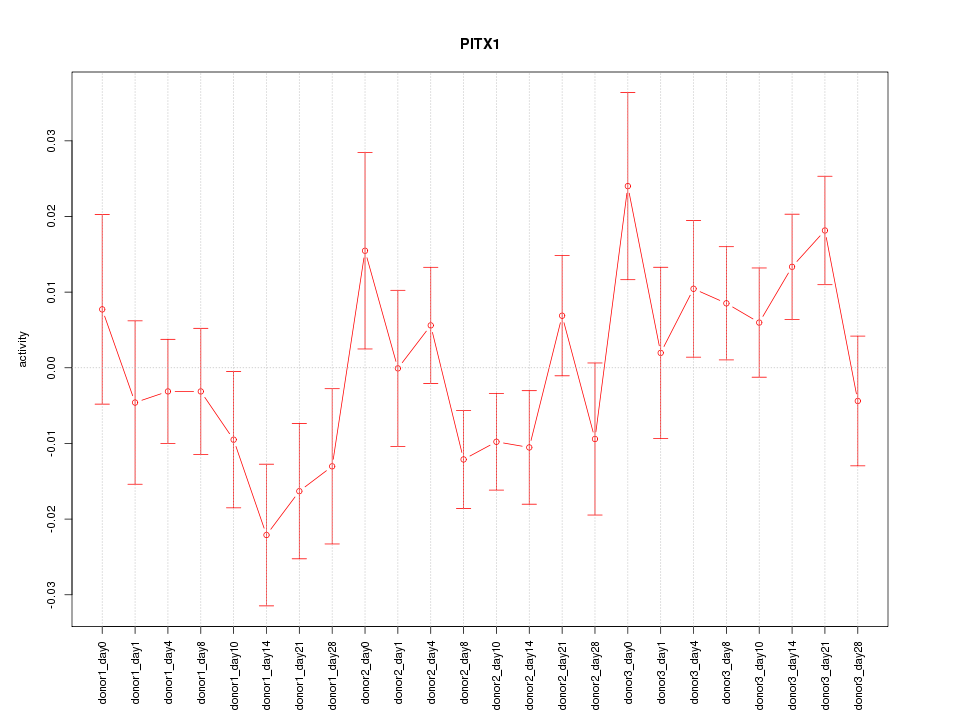
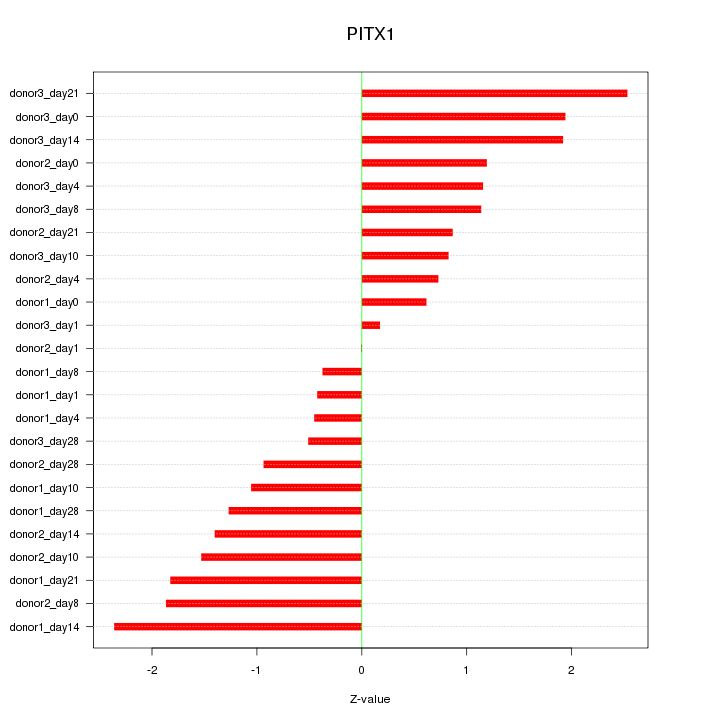
Z-value: 1.313
Transcription factors associated with PITX1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| PITX1 | ENSG00000069011.11 | PITX1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PITX1 | hg19_v2_chr5_-_134369973_134369988 | 0.43 | 3.4e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.6 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 1.1 | 3.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.9 | 2.6 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.8 | 2.4 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.8 | 3.0 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.7 | 2.0 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.6 | 1.9 | GO:2000097 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.6 | 2.4 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.6 | 5.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.5 | 4.6 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.4 | 0.4 | GO:0002784 | regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) |
| 0.4 | 1.3 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.4 | 2.5 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.4 | 1.6 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.3 | 1.0 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.3 | 1.4 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.3 | 1.0 | GO:1905166 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.3 | 1.0 | GO:1904298 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 0.3 | 1.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.3 | 1.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.3 | 0.9 | GO:2000646 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) positive regulation of receptor catabolic process(GO:2000646) |
| 0.3 | 1.5 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.3 | 0.6 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.3 | 2.3 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.3 | 1.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.3 | 1.7 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.3 | 1.6 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.3 | 5.1 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.3 | 0.8 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.3 | 1.3 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.3 | 1.5 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.2 | 1.0 | GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.2 | 1.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 0.7 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 | 3.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 1.6 | GO:0098704 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.2 | 1.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.2 | 1.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.2 | 0.6 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.2 | 3.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.2 | 0.8 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 0.8 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.2 | 2.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 1.4 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.2 | 1.0 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.2 | 0.9 | GO:0050668 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.2 | 0.7 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.2 | 1.6 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.2 | 0.5 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.2 | 0.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 0.7 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.2 | 1.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 0.7 | GO:0007159 | leukocyte cell-cell adhesion(GO:0007159) |
| 0.2 | 2.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 0.3 | GO:1901991 | negative regulation of mitotic cell cycle phase transition(GO:1901991) |
| 0.2 | 1.6 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 0.8 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.2 | 1.1 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.2 | 0.8 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.2 | 0.6 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.2 | 0.3 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.1 | 0.6 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.1 | 1.0 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.1 | 0.6 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.7 | GO:0021938 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.1 | 0.6 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 0.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 1.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.4 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.4 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.1 | 0.5 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 1.4 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 0.4 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.5 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.7 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.1 | 0.5 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.8 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.4 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.1 | 1.8 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.9 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.1 | 0.5 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.3 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 1.3 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 0.5 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 0.6 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 1.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.4 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.1 | 0.4 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.3 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.1 | 0.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.6 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.1 | 0.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.7 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 2.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.3 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.1 | 0.2 | GO:0042594 | response to starvation(GO:0042594) |
| 0.1 | 0.6 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.5 | GO:1990834 | response to odorant(GO:1990834) |
| 0.1 | 0.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.5 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.2 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.1 | 0.4 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.6 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 0.4 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.2 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.3 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.1 | 0.5 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.7 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.8 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.8 | GO:0046874 | quinolinate metabolic process(GO:0046874) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.4 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 0.3 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 0.3 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 1.4 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.2 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) aromatic amino acid family metabolic process(GO:0009072) aromatic amino acid family catabolic process(GO:0009074) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 1.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.8 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.5 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.3 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.5 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.1 | 0.7 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.3 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 | 0.2 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.1 | 0.5 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.2 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.1 | 0.8 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.1 | 0.6 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 0.5 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.1 | 0.3 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 2.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.8 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.1 | 0.3 | GO:0070981 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.1 | 0.4 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.8 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.9 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.1 | 0.4 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.2 | GO:0032888 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 0.4 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 3.1 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 0.2 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.7 | GO:0048702 | protein sulfation(GO:0006477) heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 0.8 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.1 | 0.4 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 0.6 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.1 | 0.3 | GO:0033384 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.3 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.1 | 0.6 | GO:0045141 | meiotic telomere clustering(GO:0045141) |
| 0.1 | 0.6 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.5 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.1 | 0.3 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.5 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.1 | 0.2 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.1 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.1 | 0.6 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.3 | GO:0050777 | negative regulation of immune response(GO:0050777) |
| 0.1 | 0.3 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.1 | 0.6 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 0.7 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.2 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.1 | 0.4 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.3 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 1.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.2 | GO:2001170 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.2 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 0.4 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.1 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.4 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 1.1 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.2 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 0.7 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.1 | 0.2 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.1 | 1.0 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 0.5 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 1.0 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.1 | 0.3 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.1 | 0.7 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 1.4 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.2 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) |
| 0.1 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 0.3 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.1 | 0.5 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 0.7 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 0.1 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.1 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) cellular response to water stimulus(GO:0071462) |
| 0.1 | 0.2 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.6 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.2 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) glycoprotein transport(GO:0034436) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.1 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.1 | 0.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 0.7 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.1 | 0.2 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.1 | 0.8 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.1 | 0.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.6 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 0.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.2 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.1 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.3 | GO:0051852 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 2.9 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.3 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.4 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 | 0.1 | GO:0000055 | ribosomal subunit export from nucleus(GO:0000054) ribosomal large subunit export from nucleus(GO:0000055) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.0 | 0.2 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.3 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 1.0 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.2 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.2 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 2.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 2.2 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.7 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.3 | GO:0052490 | suppression by virus of host apoptotic process(GO:0019050) negative regulation by symbiont of host apoptotic process(GO:0033668) modulation by virus of host apoptotic process(GO:0039526) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.3 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.6 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.2 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.1 | GO:2000974 | inhibition of neuroepithelial cell differentiation(GO:0002085) regulation of mitotic cell cycle, embryonic(GO:0009794) trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) mitotic cell cycle, embryonic(GO:0045448) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of timing of neuron differentiation(GO:0060164) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 1.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.4 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.2 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0010915 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 | 0.4 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.2 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.2 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.3 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.6 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.7 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 1.0 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.3 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.2 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0090024 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.3 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.5 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.2 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.2 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.0 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.3 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.1 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 2.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.0 | 0.9 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.3 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.4 | GO:0050900 | leukocyte migration(GO:0050900) |
| 0.0 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0044179 | cytolysis by symbiont of host cells(GO:0001897) hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) cytolysis in other organism involved in symbiotic interaction(GO:0051801) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.5 | GO:0051197 | positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.6 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.7 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.2 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.2 | GO:0033081 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 1.7 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 1.4 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 | 0.1 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.1 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.6 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.7 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.8 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.4 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.2 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.3 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 0.2 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.5 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.8 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 0.1 | GO:1900114 | positive regulation of histone H3-K9 dimethylation(GO:1900111) positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.3 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) |
| 0.0 | 1.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.0 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.8 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.6 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 2.3 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 0.2 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.3 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.4 | GO:0051294 | establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.3 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.4 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.5 | GO:0055082 | cellular chemical homeostasis(GO:0055082) |
| 0.0 | 0.1 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 0.5 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.3 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.6 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.2 | GO:0019441 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.5 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.0 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.1 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.3 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.0 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.4 | GO:0035909 | aorta morphogenesis(GO:0035909) |
| 0.0 | 0.4 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.8 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.5 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.3 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.4 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.0 | 0.5 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.3 | GO:0023058 | adaptation of signaling pathway(GO:0023058) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0071352 | interleukin-2-mediated signaling pathway(GO:0038110) response to interleukin-2(GO:0070669) cellular response to interleukin-2(GO:0071352) |
| 0.0 | 0.1 | GO:0060312 | regulation of blood vessel remodeling(GO:0060312) |
| 0.0 | 0.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.0 | 0.2 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 0.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0018194 | N-terminal protein amino acid methylation(GO:0006480) N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.0 | 0.2 | GO:1903027 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 1.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 1.4 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) |
| 0.0 | 0.3 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.1 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 0.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 1.0 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0070586 | cell migration involved in gastrulation(GO:0042074) cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.0 | 0.4 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.5 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.3 | GO:0034694 | response to prostaglandin(GO:0034694) |
| 0.0 | 0.1 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.4 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.2 | GO:0007077 | mitotic nuclear envelope disassembly(GO:0007077) |
| 0.0 | 0.9 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.3 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.2 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.5 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.0 | 0.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.0 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.1 | GO:0032740 | positive regulation of interleukin-17 production(GO:0032740) |
| 0.0 | 0.2 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.1 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.2 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.3 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.0 | 0.0 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.3 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.0 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.3 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.4 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.7 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.5 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.0 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:1903522 | regulation of blood circulation(GO:1903522) |
| 0.0 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.2 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.3 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.7 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.2 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.6 | 2.4 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.4 | 1.8 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.3 | 2.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.3 | 2.5 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.3 | 0.9 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.3 | 0.8 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.3 | 1.0 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.2 | 5.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 0.6 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 3.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.5 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 0.5 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.8 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.6 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.1 | 0.5 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 1.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 1.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.3 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.1 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 1.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.4 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 0.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.2 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.3 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 1.0 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 0.2 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.1 | 0.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 1.0 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 2.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.7 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 1.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.4 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 0.6 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.5 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 1.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.2 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 1.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.7 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.9 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.2 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.4 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 2.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.7 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 1.9 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.7 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 1.1 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.6 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.5 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.7 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.0 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 3.1 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.1 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.5 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 1.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 2.8 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0044853 | plasma membrane raft(GO:0044853) |
| 0.0 | 0.4 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.7 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 2.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.9 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 5.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.4 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 7.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.6 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 2.3 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.1 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 3.0 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.6 | 2.4 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.6 | 3.0 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.5 | 4.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.5 | 3.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.5 | 1.4 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.4 | 1.7 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.4 | 1.6 | GO:0047718 | indanol dehydrogenase activity(GO:0047718) |
| 0.4 | 1.2 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.4 | 5.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.3 | 1.0 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.3 | 0.6 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.3 | 2.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.3 | 0.8 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.3 | 0.8 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.3 | 1.0 | GO:0016296 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.3 | 0.8 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.3 | 2.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 0.7 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.2 | 0.9 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.2 | 1.6 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.2 | 2.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.2 | 0.8 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.2 | 0.8 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.2 | 0.8 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 1.0 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.2 | 1.0 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 0.8 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.2 | 0.8 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.2 | 1.9 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 0.6 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.2 | 0.7 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.2 | 0.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 0.5 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.2 | 1.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.2 | 1.0 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.2 | 0.5 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.2 | 2.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 1.0 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 1.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 0.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 2.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 0.8 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.2 | 0.8 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.2 | 3.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 0.8 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 1.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.7 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.1 | 1.1 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.6 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.7 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 0.7 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.7 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.8 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 1.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.6 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 1.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.4 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 0.5 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 1.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 0.6 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 1.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 2.7 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.1 | 0.5 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 0.1 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.1 | 0.3 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.1 | 3.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.6 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.1 | 0.4 | GO:0030395 | lactose binding(GO:0030395) |
| 0.1 | 0.4 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 0.3 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.9 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.3 | GO:0016517 | interleukin-12 receptor activity(GO:0016517) |
| 0.1 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 0.4 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.5 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.8 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) purinergic receptor activity(GO:0035586) |
| 0.1 | 1.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.9 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.1 | 0.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.8 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.5 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 11.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.3 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.1 | 1.0 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.2 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 1.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.5 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.2 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.2 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 1.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.3 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.3 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.1 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 2.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.3 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 1.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.4 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 2.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.3 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 0.2 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.1 | 0.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.5 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.2 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.1 | 0.3 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.2 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.1 | 0.2 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 1.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 0.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.3 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.1 | 0.5 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.3 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 0.6 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.7 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 0.3 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 7.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.2 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.2 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 0.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.3 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 1.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.2 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 2.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 0.6 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.6 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0005230 | extracellular ligand-gated ion channel activity(GO:0005230) |
| 0.0 | 0.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.3 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.2 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 1.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.4 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 2.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0070290 | phospholipase D activity(GO:0004630) N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.3 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.3 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 1.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.5 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0004917 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.2 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 2.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 1.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.0 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 2.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.8 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.0 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.3 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.8 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 5.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.0 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 1.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0070840 | dynein complex binding(GO:0070840) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | ST_INTERLEUKIN_4_PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 3.9 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.4 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 4.9 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.1 | 3.8 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.9 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.3 | PID_S1P_S1P1_PATHWAY | S1P1 pathway |
| 0.1 | 0.3 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 2.9 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 3.3 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.2 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.0 | PID_CXCR3_PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.5 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 2.8 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.3 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID_ECADHERIN_KERATINOCYTE_PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID_IL27_PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.1 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.6 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.0 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.2 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.8 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 8.5 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.5 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.2 | PID_TCR_PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.3 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.8 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.1 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.1 | PID_S1P_S1P3_PATHWAY | S1P3 pathway |
| 0.0 | 0.4 | ST_GA13_PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.2 | PID_PI3KCI_AKT_PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 4.7 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID_PI3K_PLC_TRK_PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.4 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | PID_AVB3_OPN_PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.2 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.6 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.0 | 2.7 | NABA_MATRISOME_ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.2 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.6 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 3.4 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.6 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 4.0 | REACTOME_RIP_MEDIATED_NFKB_ACTIVATION_VIA_DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 0.6 | REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.3 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.1 | REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.0 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 3.9 | REACTOME_PTM_GAMMA_CARBOXYLATION_HYPUSINE_FORMATION_AND_ARYLSULFATASE_ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 1.2 | REACTOME_OPSINS | Genes involved in Opsins |
| 0.1 | 1.8 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.6 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 5.6 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.4 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.0 | REACTOME_ANDROGEN_BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.1 | REACTOME_REVERSIBLE_HYDRATION_OF_CARBON_DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.4 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.2 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.0 | REACTOME_ENDOGENOUS_STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 2.3 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.0 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.1 | 2.0 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.3 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.9 | REACTOME_POST_TRANSLATIONAL_MODIFICATION_SYNTHESIS_OF_GPI_ANCHORED_PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.3 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME_ADP_SIGNALLING_THROUGH_P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 2.4 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME_TRAFFICKING_AND_PROCESSING_OF_ENDOSOMAL_TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.7 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.8 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.8 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.8 | REACTOME_HIGHLY_CALCIUM_PERMEABLE_POSTSYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.5 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.9 | REACTOME_PYRUVATE_METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.7 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME_TAK1_ACTIVATES_NFKB_BY_PHOSPHORYLATION_AND_ACTIVATION_OF_IKKS_COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 2.0 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.0 | REACTOME_PROSTACYCLIN_SIGNALLING_THROUGH_PROSTACYCLIN_RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 1.3 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.2 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.9 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.9 | REACTOME_ACTIVATION_OF_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 1.1 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.0 | 0.7 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.7 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.8 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.6 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.8 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.6 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 3.3 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME_THROMBOXANE_SIGNALLING_THROUGH_TP_RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.3 | REACTOME_CD28_DEPENDENT_VAV1_PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 5.1 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.7 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.5 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.9 | REACTOME_REGULATION_OF_HYPOXIA_INDUCIBLE_FACTOR_HIF_BY_OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.6 | REACTOME_HOST_INTERACTIONS_OF_HIV_FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.8 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.8 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME_TRYPTOPHAN_CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.6 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.7 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME_CTLA4_INHIBITORY_SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.7 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.2 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.1 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME_RESOLUTION_OF_AP_SITES_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME_SIGNAL_TRANSDUCTION_BY_L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.2 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.3 | REACTOME_APC_C_CDH1_MEDIATED_DEGRADATION_OF_CDC20_AND_OTHER_APC_C_CDH1_TARGETED_PROTEINS_IN_LATE_MITOSIS_EARLY_G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.6 | REACTOME_SRP_DEPENDENT_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.1 | REACTOME_G_ALPHA_S_SIGNALLING_EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.1 | REACTOME_INFLUENZA_VIRAL_RNA_TRANSCRIPTION_AND_REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.2 | REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.5 | REACTOME_G1_PHASE | Genes involved in G1 Phase |