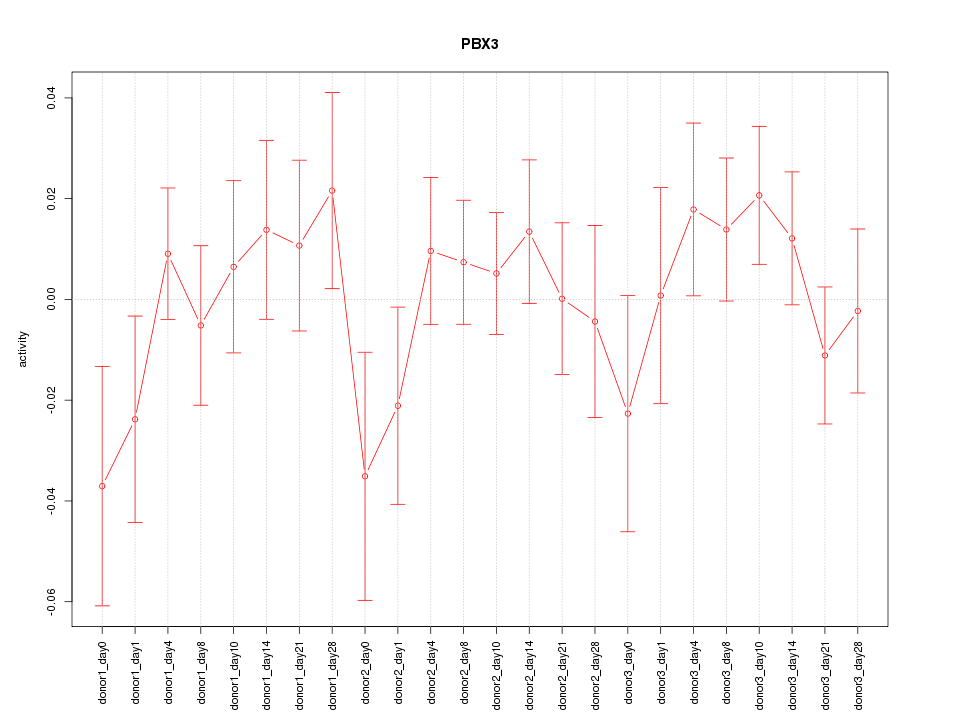
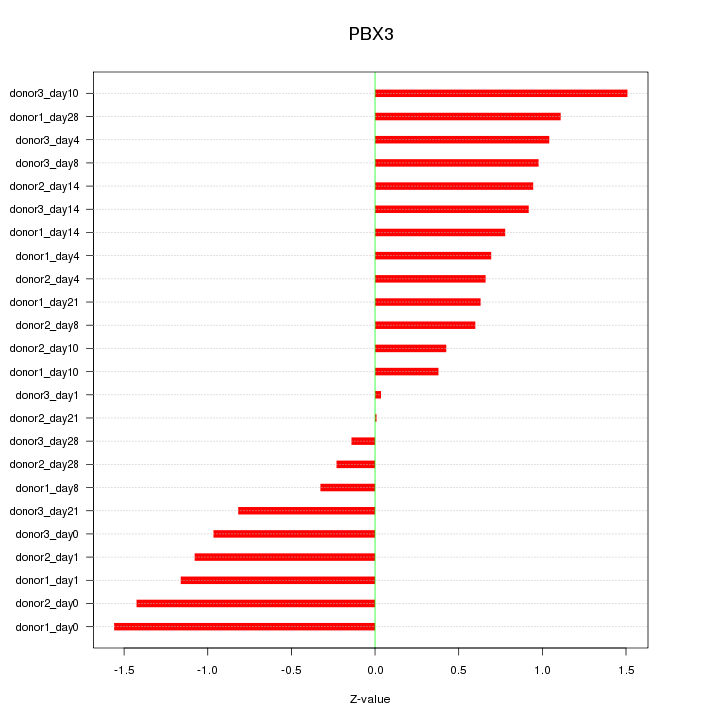
Motif ID: PBX3
Z-value: 0.881
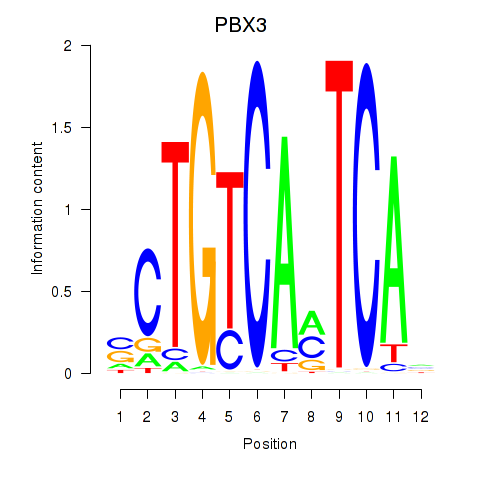
Transcription factors associated with PBX3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| PBX3 | ENSG00000167081.12 | PBX3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PBX3 | hg19_v2_chr9_+_128509663_128509733 | -0.67 | 3.1e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.3 | 1.0 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.3 | 1.5 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.3 | 1.7 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.3 | 0.8 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.3 | 0.8 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.2 | 0.7 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.2 | 1.5 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.2 | 0.7 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.2 | 1.3 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.2 | 1.1 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 0.8 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 | 0.6 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.2 | 0.5 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 | 0.5 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.2 | 0.6 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 0.9 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.1 | 0.9 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.6 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 | 0.1 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.1 | 1.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.1 | 0.4 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.4 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.5 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.3 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.3 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.1 | 0.5 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 0.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 1.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.7 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.2 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.5 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.4 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.1 | 0.3 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.1 | 0.3 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 0.3 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 1.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.7 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.1 | 0.3 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 1.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.6 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.3 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.1 | 0.9 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.3 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.2 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.4 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.3 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.3 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 0.4 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.1 | 0.2 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 | 1.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.3 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.3 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.1 | 0.4 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 1.9 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.1 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.8 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.5 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.1 | 0.5 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.3 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.1 | 0.4 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 0.7 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.2 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.1 | 0.3 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 1.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.3 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.7 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.3 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 2.0 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.2 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.3 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 1.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.8 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 0.1 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.9 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.3 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:2000617 | positive regulation of histone H3-K9 acetylation(GO:2000617) |
| 0.0 | 1.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 2.1 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 1.0 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.4 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 1.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.1 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.7 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.6 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.2 | GO:0046465 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.1 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.1 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.1 | GO:0044854 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.0 | 0.5 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.2 | GO:0048243 | norepinephrine secretion(GO:0048243) |
| 0.0 | 0.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.3 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 2.2 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 0.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.4 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.3 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.5 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.8 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.0 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.0 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.3 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.0 | 0.1 | GO:0060325 | face development(GO:0060324) face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.5 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.1 | GO:0032909 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0001534 | radial spoke(GO:0001534) |
| 0.2 | 0.7 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.9 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.9 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.5 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.5 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 1.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 1.7 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.8 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.5 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 1.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.0 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.5 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.4 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 1.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.4 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.0 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.6 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 1.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 2.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.7 | GO:0005657 | replication fork(GO:0005657) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.5 | 1.5 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.3 | 0.9 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.3 | 1.5 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.3 | 1.0 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.2 | 1.7 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.2 | 0.8 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.2 | 1.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.2 | 1.2 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 1.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 1.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 1.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.8 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 0.4 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 1.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.3 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 1.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.4 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 1.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.3 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.3 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 0.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.5 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.7 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.5 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.6 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.1 | 0.9 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.2 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 1.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.2 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.1 | 1.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 0.6 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 1.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 1.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 4.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.0 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.3 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 2.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.9 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0016462 | pyrophosphatase activity(GO:0016462) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 1.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.2 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.9 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.2 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.0 | 1.5 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.5 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.4 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.4 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.5 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.5 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 4.3 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.0 | REACTOME_A_TETRASACCHARIDE_LINKER_SEQUENCE_IS_REQUIRED_FOR_GAG_SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.0 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.0 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.7 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.1 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.5 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.1 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.0 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.9 | REACTOME_PEROXISOMAL_LIPID_METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 8.8 | REACTOME_GENERIC_TRANSCRIPTION_PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.3 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME_PLATELET_SENSITIZATION_BY_LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.4 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.4 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.5 | REACTOME_PERK_REGULATED_GENE_EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.4 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.1 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME_ACTIVATION_OF_NF_KAPPAB_IN_B_CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.5 | REACTOME_ERK_MAPK_TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.2 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME_CIRCADIAN_CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.5 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |