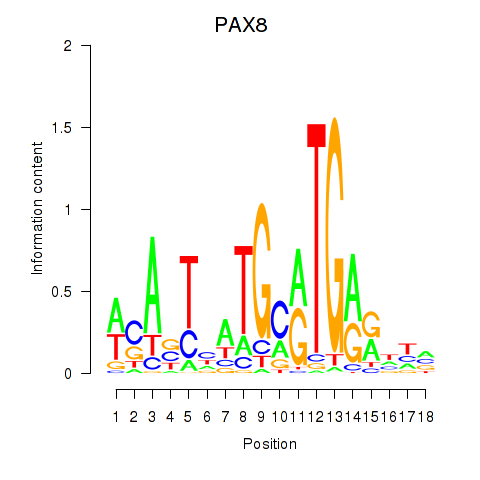
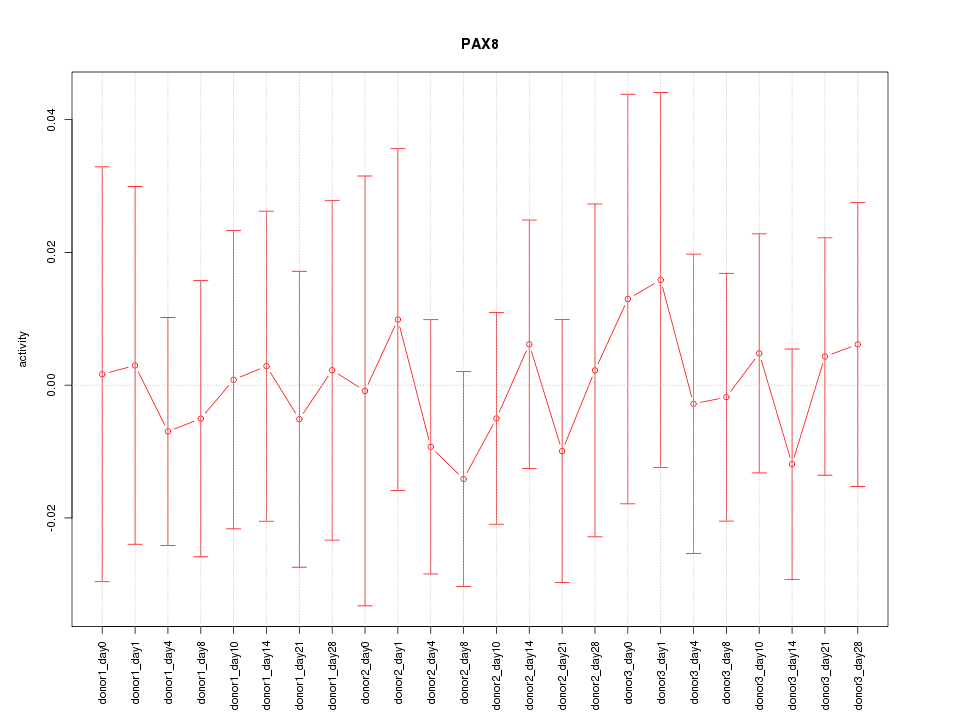
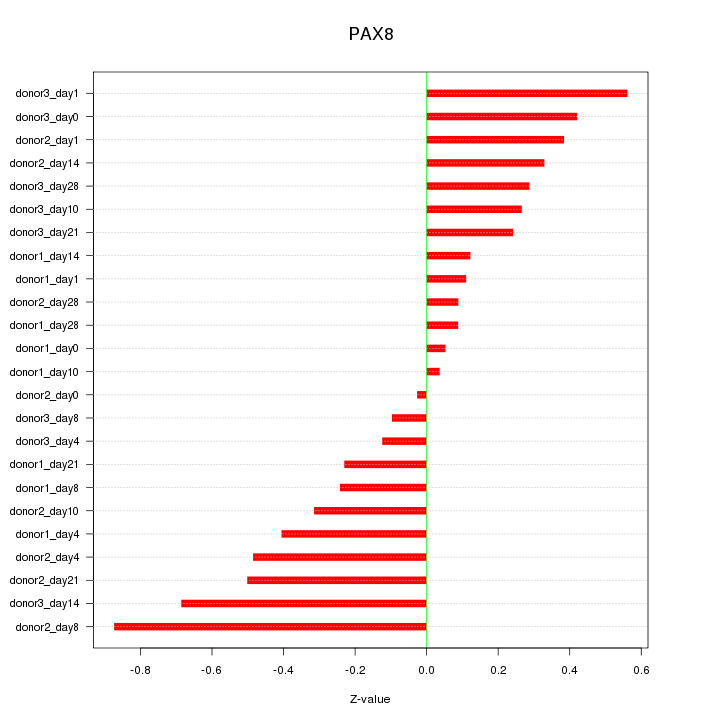
| 0.2 |
0.5 |
GO:0042700 |
luteinizing hormone signaling pathway(GO:0042700) |
| 0.1 |
0.7 |
GO:0002803 |
positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 |
0.2 |
GO:0002644 |
negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 |
0.3 |
GO:0070981 |
L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.1 |
0.2 |
GO:0010752 |
regulation of cGMP-mediated signaling(GO:0010752) |
| 0.1 |
0.2 |
GO:0002581 |
negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 |
0.2 |
GO:1903570 |
regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 |
0.3 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.1 |
GO:0046005 |
positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 |
0.1 |
GO:0090271 |
positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 |
0.3 |
GO:0010193 |
response to ozone(GO:0010193) |
| 0.0 |
0.3 |
GO:0035803 |
egg coat formation(GO:0035803) |
| 0.0 |
0.3 |
GO:0050859 |
negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 |
0.1 |
GO:0072709 |
cellular response to sorbitol(GO:0072709) |
| 0.0 |
0.3 |
GO:1903575 |
cornified envelope assembly(GO:1903575) |
| 0.0 |
0.1 |
GO:2000393 |
negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 |
0.2 |
GO:0061767 |
negative regulation of lung blood pressure(GO:0061767) |
| 0.0 |
0.1 |
GO:0036292 |
DNA rewinding(GO:0036292) |
| 0.0 |
0.1 |
GO:0035990 |
tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 |
0.2 |
GO:0021842 |
directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 |
0.2 |
GO:0038060 |
nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 |
0.2 |
GO:0048241 |
epinephrine transport(GO:0048241) |
| 0.0 |
0.1 |
GO:2000523 |
regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 |
0.2 |
GO:2000252 |
negative regulation of feeding behavior(GO:2000252) |
| 0.0 |
0.1 |
GO:0050960 |
detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 |
0.1 |
GO:0009822 |
alkaloid catabolic process(GO:0009822) |
| 0.0 |
0.1 |
GO:0046167 |
glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 |
0.3 |
GO:0045625 |
regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 |
0.1 |
GO:0007500 |
mesodermal cell fate determination(GO:0007500) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 |
0.1 |
GO:0042357 |
thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 |
0.1 |
GO:0090131 |
mesenchyme migration(GO:0090131) |
| 0.0 |
0.3 |
GO:0098828 |
positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 |
0.2 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.0 |
0.1 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 |
2.0 |
GO:0070268 |
cornification(GO:0070268) |
| 0.0 |
0.1 |
GO:0071874 |
cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 |
0.1 |
GO:0072233 |
thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 |
0.1 |
GO:0046968 |
peptide antigen transport(GO:0046968) |
| 0.0 |
0.0 |
GO:0032641 |
lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 |
0.1 |
GO:2000402 |
negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 |
0.2 |
GO:2000773 |
negative regulation of cellular senescence(GO:2000773) |
| 0.0 |
0.1 |
GO:0010993 |
regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 |
0.1 |
GO:0010734 |
protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 |
0.1 |
GO:0019441 |
tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 |
0.1 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
0.1 |
GO:0030950 |
establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 |
0.2 |
GO:0033141 |
positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 |
0.4 |
GO:0034080 |
CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 |
0.1 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.0 |
0.1 |
GO:2001206 |
positive regulation of osteoclast development(GO:2001206) |
| 0.0 |
0.1 |
GO:2001300 |
lipoxin metabolic process(GO:2001300) |
| 0.0 |
0.1 |
GO:0006857 |
oligopeptide transport(GO:0006857) |