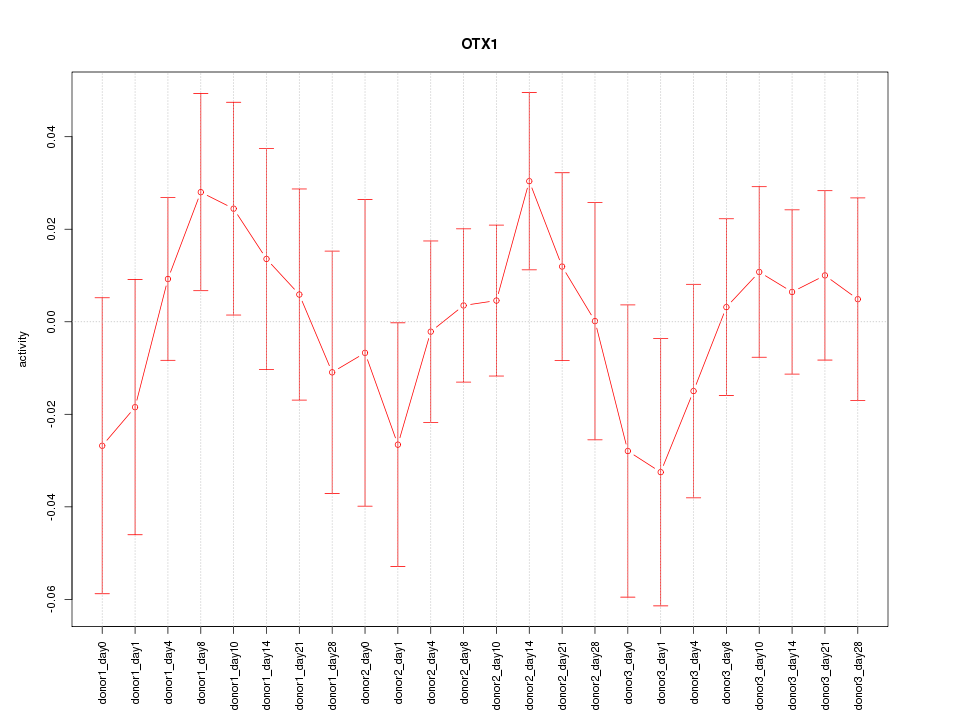
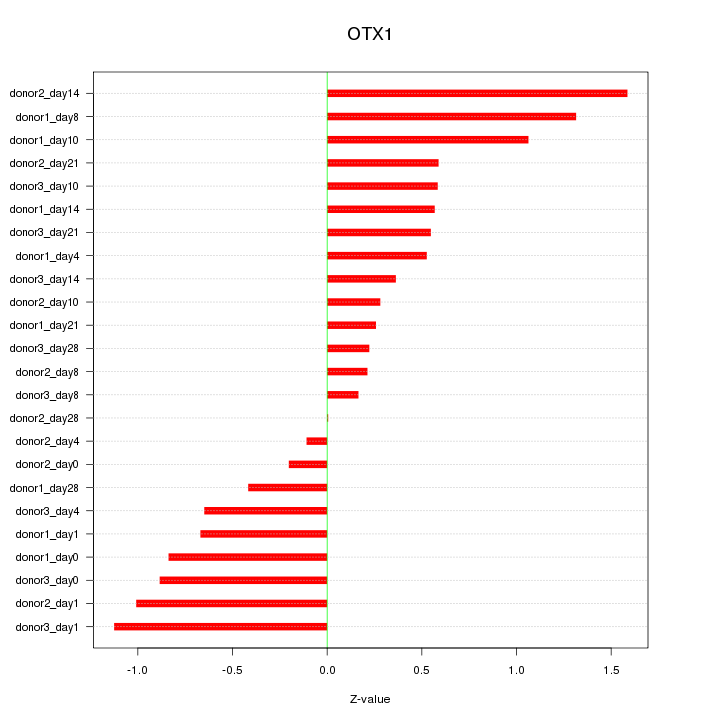
Motif ID: OTX1
Z-value: 0.714
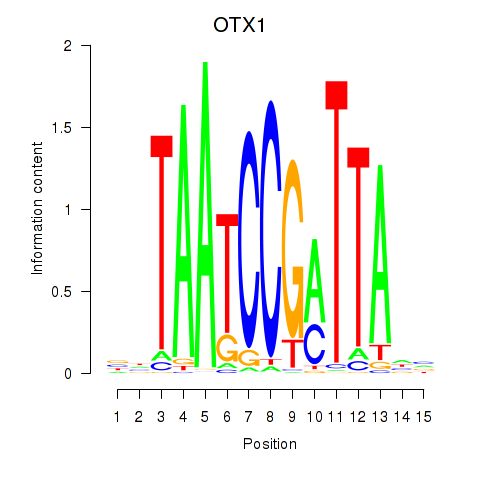
Transcription factors associated with OTX1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| OTX1 | ENSG00000115507.5 | OTX1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OTX1 | hg19_v2_chr2_+_63277927_63277938 | 0.57 | 3.3e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.3 | 8.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 1.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 1.4 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 0.5 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.7 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.3 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.3 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.3 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 0.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.3 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 1.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 3.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.2 | GO:2000653 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.1 | 0.2 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.5 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.3 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.7 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 1.2 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.9 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.5 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.3 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 2.3 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.1 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.1 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.4 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 1.3 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.6 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.4 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.0 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.4 | GO:0006511 | ubiquitin-dependent protein catabolic process(GO:0006511) modification-dependent protein catabolic process(GO:0019941) modification-dependent macromolecule catabolic process(GO:0043632) |
| 0.0 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 1.0 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.6 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 1.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.6 | GO:1902911 | protein kinase complex(GO:1902911) |
| 0.0 | 0.4 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 2.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 1.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 1.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.2 | 1.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 3.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 1.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.4 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 1.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 1.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.0 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.0 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.3 | ST_ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.7 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.9 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | REACTOME_RECYCLING_OF_BILE_ACIDS_AND_SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.2 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.5 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.1 | REACTOME_GLUCAGON_TYPE_LIGAND_RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.6 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME_TETRAHYDROBIOPTERIN_BH4_SYNTHESIS_RECYCLING_SALVAGE_AND_REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.5 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.0 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |