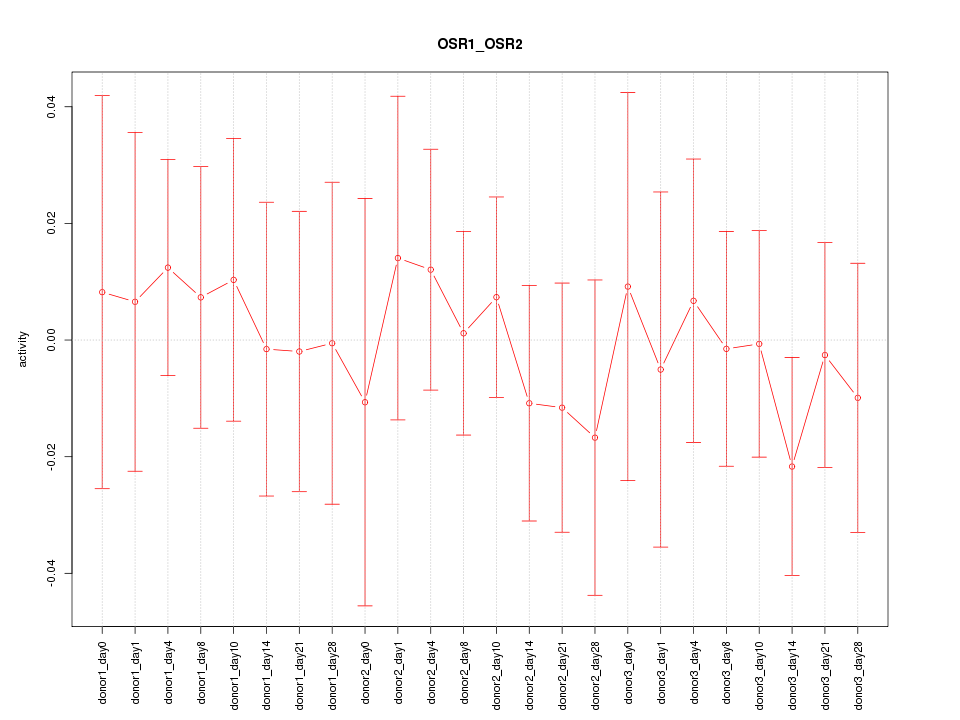
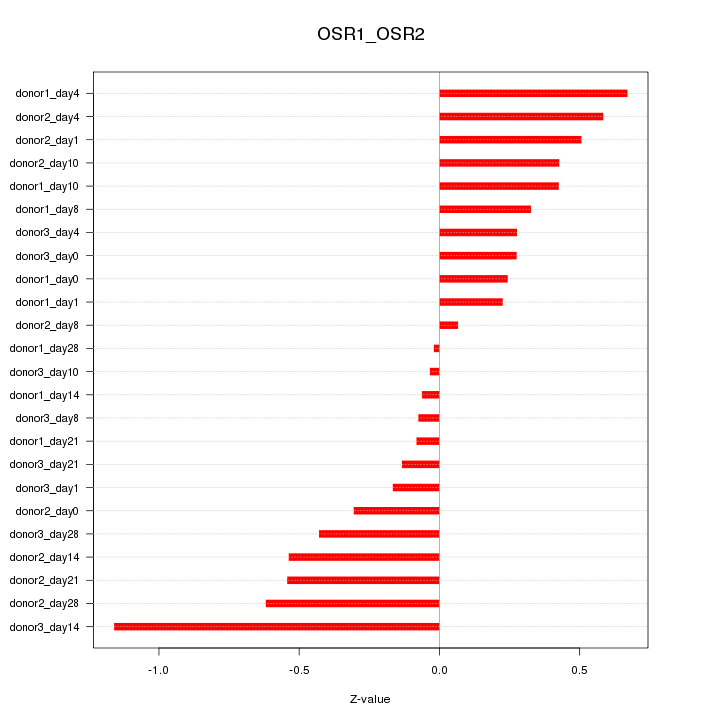
Motif ID: OSR1_OSR2
Z-value: 0.430
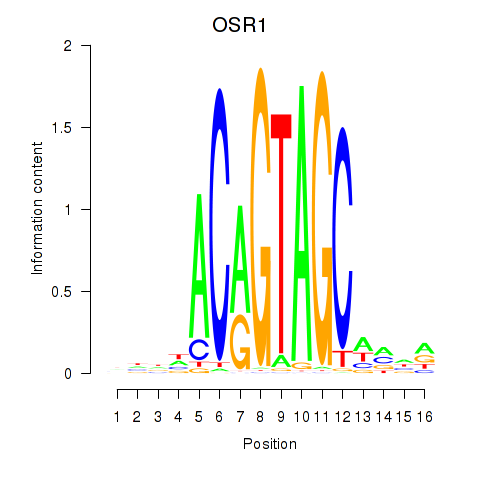
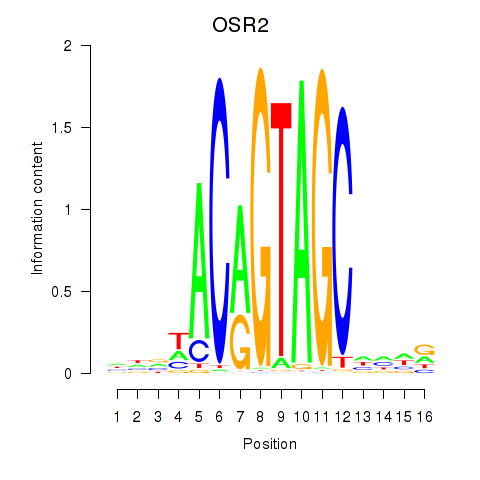
Transcription factors associated with OSR1_OSR2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| OSR1 | ENSG00000143867.5 | OSR1 |
| OSR2 | ENSG00000164920.5 | OSR2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OSR2 | hg19_v2_chr8_+_99956759_99956887 | -0.33 | 1.2e-01 | Click! |
| OSR1 | hg19_v2_chr2_-_19558373_19558414 | -0.13 | 5.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.1 | 0.6 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.5 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.3 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.1 | 0.3 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 0.2 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.0 | 0.2 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.0 | 0.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.2 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.1 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.6 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.0 | GO:1903423 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.0 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.0 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0030805 | regulation of cyclic nucleotide catabolic process(GO:0030805) regulation of cAMP catabolic process(GO:0030820) regulation of purine nucleotide catabolic process(GO:0033121) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.0 | 0.1 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.6 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.3 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.6 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.0 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.5 | GO:0047718 | indanol dehydrogenase activity(GO:0047718) |
| 0.1 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0070283 | lipoate synthase activity(GO:0016992) radical SAM enzyme activity(GO:0070283) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID_PLK1_PATHWAY | PLK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME_E2F_ENABLED_INHIBITION_OF_PRE_REPLICATION_COMPLEX_FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.6 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |