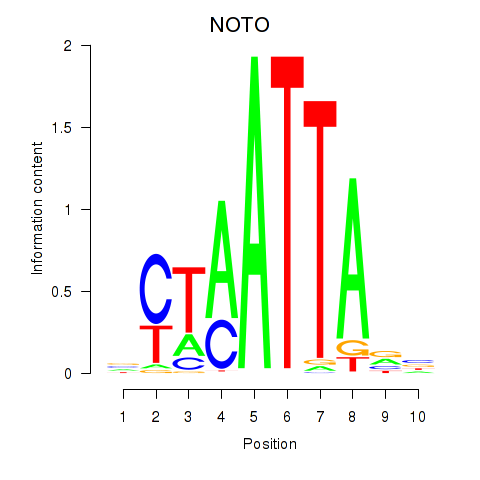
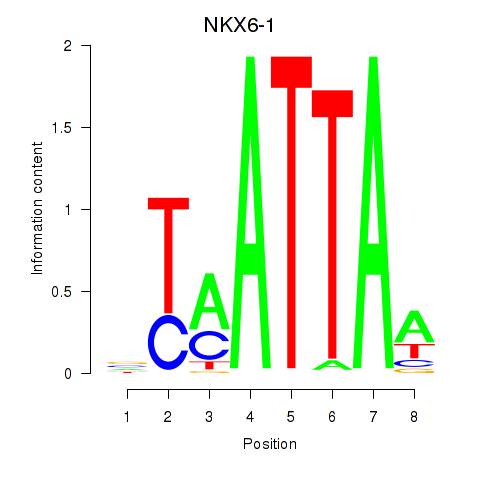
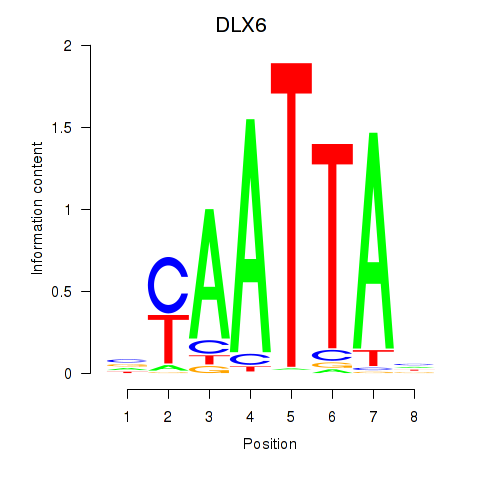
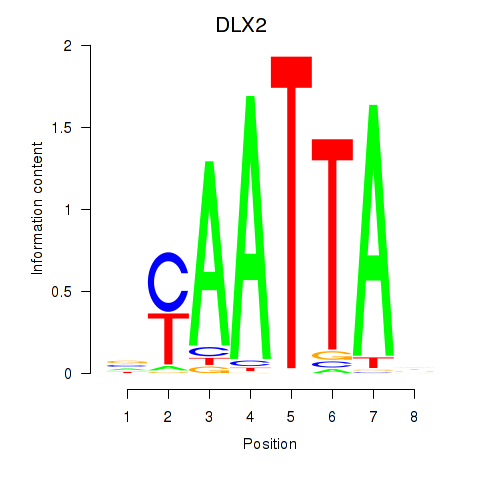
| 0.3 |
0.9 |
GO:1905072 |
apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.2 |
0.7 |
GO:0003147 |
neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.2 |
1.4 |
GO:2000324 |
positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.2 |
1.9 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 |
0.6 |
GO:0070173 |
regulation of enamel mineralization(GO:0070173) |
| 0.2 |
1.1 |
GO:1902748 |
positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 |
1.9 |
GO:0032277 |
negative regulation of gonadotropin secretion(GO:0032277) |
| 0.2 |
0.3 |
GO:2000854 |
positive regulation of corticosterone secretion(GO:2000854) |
| 0.2 |
0.5 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 |
0.8 |
GO:0048023 |
positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 |
2.2 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.1 |
0.4 |
GO:1904899 |
regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 |
1.8 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.1 |
1.5 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.1 |
0.9 |
GO:0006627 |
protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 |
0.1 |
GO:0060738 |
epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.1 |
0.2 |
GO:1902725 |
negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 |
0.4 |
GO:0072658 |
maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 |
0.3 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.1 |
0.3 |
GO:1904580 |
regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 |
0.9 |
GO:0021520 |
spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 |
0.2 |
GO:0099543 |
retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 |
1.6 |
GO:0072189 |
ureter development(GO:0072189) |
| 0.1 |
0.2 |
GO:0098582 |
innate vocalization behavior(GO:0098582) |
| 0.1 |
0.2 |
GO:2001190 |
positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 |
1.0 |
GO:0002862 |
negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 |
0.2 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 |
0.6 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.1 |
0.5 |
GO:0060287 |
epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 |
0.7 |
GO:0006228 |
UTP biosynthetic process(GO:0006228) |
| 0.1 |
1.3 |
GO:0006068 |
ethanol catabolic process(GO:0006068) |
| 0.1 |
1.0 |
GO:0016024 |
CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 |
0.5 |
GO:0051388 |
positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 |
0.2 |
GO:0003172 |
primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 |
0.5 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.0 |
0.2 |
GO:0031296 |
positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 |
1.2 |
GO:0097067 |
cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 |
0.3 |
GO:0051970 |
negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 |
0.7 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.0 |
0.2 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.0 |
0.1 |
GO:1904640 |
response to methionine(GO:1904640) |
| 0.0 |
0.6 |
GO:0046600 |
negative regulation of centriole replication(GO:0046600) |
| 0.0 |
0.2 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 |
0.2 |
GO:1904383 |
response to sodium phosphate(GO:1904383) |
| 0.0 |
1.9 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.4 |
GO:0034214 |
protein hexamerization(GO:0034214) |
| 0.0 |
1.9 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.0 |
0.2 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.0 |
0.3 |
GO:0071461 |
cellular response to redox state(GO:0071461) |
| 0.0 |
0.4 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 |
0.2 |
GO:0052199 |
negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 |
0.2 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 |
0.2 |
GO:0038161 |
prolactin signaling pathway(GO:0038161) |
| 0.0 |
0.2 |
GO:0015692 |
lead ion transport(GO:0015692) |
| 0.0 |
0.2 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 |
0.1 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.0 |
0.3 |
GO:0002414 |
immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 |
0.5 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
0.0 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.0 |
0.1 |
GO:0051958 |
methotrexate transport(GO:0051958) |
| 0.0 |
0.3 |
GO:0045078 |
positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 |
0.2 |
GO:0015842 |
aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 |
0.4 |
GO:0070257 |
positive regulation of mucus secretion(GO:0070257) |
| 0.0 |
0.2 |
GO:0060437 |
lung growth(GO:0060437) |
| 0.0 |
0.1 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) |
| 0.0 |
0.1 |
GO:1904245 |
regulation of polynucleotide adenylyltransferase activity(GO:1904245) positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 |
0.1 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 |
0.1 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 |
0.1 |
GO:0010760 |
negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 |
0.1 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.1 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 |
0.1 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 |
0.2 |
GO:0060020 |
Bergmann glial cell differentiation(GO:0060020) |
| 0.0 |
0.3 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.0 |
0.6 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.0 |
1.0 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.2 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.0 |
0.1 |
GO:0070105 |
positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 |
0.1 |
GO:0002018 |
renin-angiotensin regulation of aldosterone production(GO:0002018) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 |
0.1 |
GO:0070309 |
lens fiber cell morphogenesis(GO:0070309) |
| 0.0 |
0.3 |
GO:0060397 |
JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 |
0.1 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 |
0.1 |
GO:0031860 |
telomeric 3' overhang formation(GO:0031860) |
| 0.0 |
0.1 |
GO:0044245 |
polysaccharide digestion(GO:0044245) |
| 0.0 |
0.0 |
GO:0060355 |
positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 |
0.1 |
GO:0046005 |
positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 |
0.3 |
GO:0031647 |
regulation of protein stability(GO:0031647) |
| 0.0 |
0.0 |
GO:0038188 |
cholecystokinin signaling pathway(GO:0038188) |
| 0.0 |
0.4 |
GO:0044458 |
motile cilium assembly(GO:0044458) |
| 0.0 |
0.1 |
GO:0010748 |
regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 |
0.0 |
GO:0021730 |
trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.0 |
0.5 |
GO:0021680 |
cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 |
0.1 |
GO:0050916 |
sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 |
0.1 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.1 |
GO:0010157 |
response to chlorate(GO:0010157) |
| 0.0 |
0.1 |
GO:0071638 |
adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 |
0.1 |
GO:0006196 |
AMP catabolic process(GO:0006196) |
| 0.0 |
0.9 |
GO:0007595 |
lactation(GO:0007595) |
| 0.0 |
0.1 |
GO:0060012 |
synaptic transmission, glycinergic(GO:0060012) |
| 0.0 |
0.1 |
GO:0042492 |
gamma-delta T cell differentiation(GO:0042492) |
| 0.0 |
0.2 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 |
0.4 |
GO:0031954 |
positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 |
0.1 |
GO:0086024 |
adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 |
0.5 |
GO:2000144 |
positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 |
0.1 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.0 |
0.1 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.0 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 |
0.4 |
GO:0001919 |
regulation of receptor recycling(GO:0001919) |
| 0.0 |
0.2 |
GO:0035372 |
protein localization to microtubule(GO:0035372) |
| 0.0 |
0.0 |
GO:0009956 |
radial pattern formation(GO:0009956) |
| 0.0 |
0.0 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 |
0.1 |
GO:0061669 |
spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 |
0.5 |
GO:0007274 |
neuromuscular synaptic transmission(GO:0007274) |
| 0.0 |
0.1 |
GO:0060414 |
aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 |
0.2 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.1 |
GO:0002074 |
extraocular skeletal muscle development(GO:0002074) |
| 0.0 |
0.1 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.9 |
GO:0007585 |
respiratory gaseous exchange(GO:0007585) |
| 0.0 |
0.2 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.1 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |