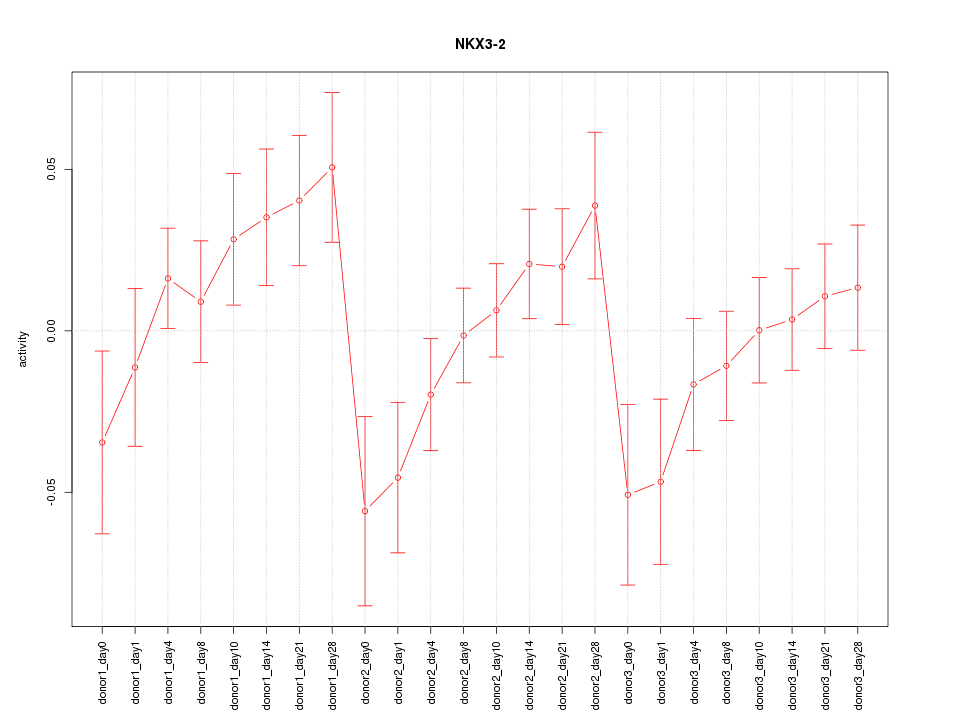
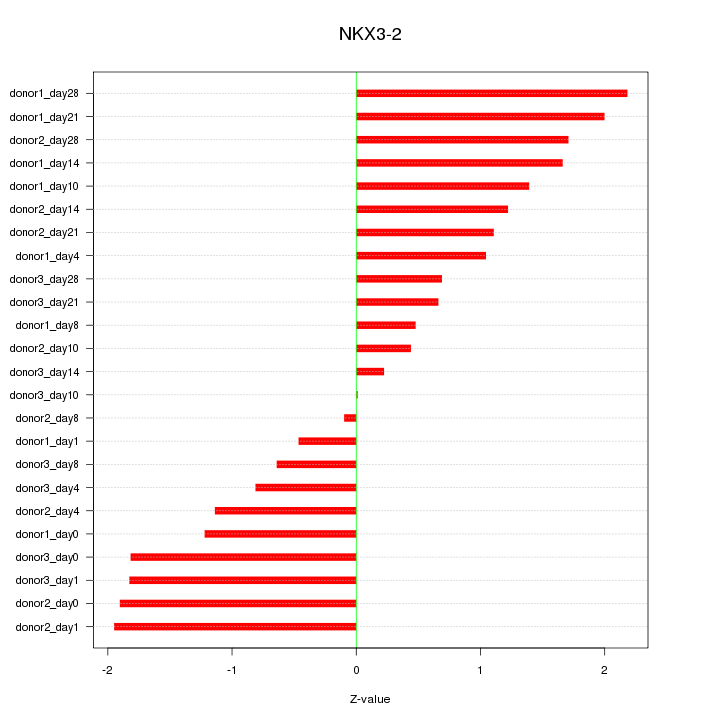
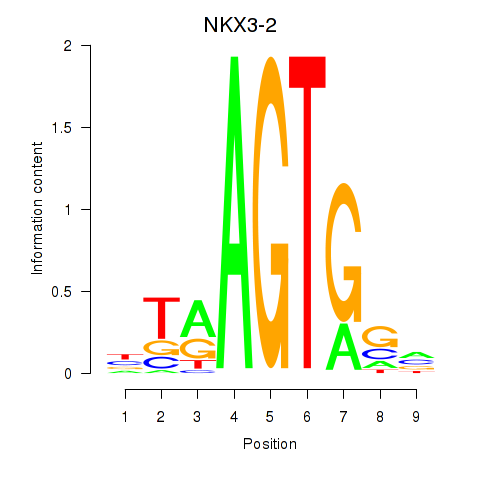
Motif ID: NKX3-2
Z-value: 1.286
Transcription factors associated with NKX3-2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NKX3-2 | ENSG00000109705.7 | NKX3-2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX3-2 | hg19_v2_chr4_-_13546632_13546674 | -0.25 | 2.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 1.2 | 6.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 1.1 | 5.7 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 1.1 | 3.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.8 | 2.4 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.8 | 3.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.6 | 6.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.4 | 1.3 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.4 | 5.3 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.4 | 8.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 | 1.1 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.3 | 1.2 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.3 | 0.8 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.3 | 3.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.3 | 5.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.3 | 0.8 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.3 | 1.0 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.3 | 1.3 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.2 | 0.7 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.2 | 0.7 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.2 | 1.7 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.2 | 2.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 1.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 3.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.2 | 0.9 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.2 | 2.5 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.2 | 0.7 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 1.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 6.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 0.3 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.2 | 0.8 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 1.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.1 | 0.6 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 0.7 | GO:1900920 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 | 1.3 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 2.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 1.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.7 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.1 | 9.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.4 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 0.3 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 6.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 5.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.6 | GO:2000302 | regulation of synaptic vesicle priming(GO:0010807) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.1 | 0.3 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 1.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.3 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.3 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 1.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.8 | GO:2000546 | positive regulation of fat cell proliferation(GO:0070346) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.3 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 1.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 1.0 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 2.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 7.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.4 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.1 | 0.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.4 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 2.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 1.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.9 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 1.9 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 2.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.4 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.1 | 0.7 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.1 | 0.5 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 2.8 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.1 | 0.8 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.3 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.1 | 0.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.7 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 0.3 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 0.9 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 1.0 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.5 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.5 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.7 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 1.2 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 1.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 5.7 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 1.0 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 1.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.3 | GO:0008037 | cell recognition(GO:0008037) |
| 0.0 | 0.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.4 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 0.2 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.0 | 0.9 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.8 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 2.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.7 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.3 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.3 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.1 | GO:2000861 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) positive regulation of estrogen secretion(GO:2000863) regulation of estradiol secretion(GO:2000864) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 1.7 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.4 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 3.5 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 1.2 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.0 | 0.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.3 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.3 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 1.2 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.3 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.3 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.9 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.5 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.1 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.0 | 1.0 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.8 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.7 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 1.0 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.6 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.2 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0001505 | regulation of neurotransmitter levels(GO:0001505) |
| 0.0 | 0.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0051934 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.4 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.6 | 2.8 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.5 | 5.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.4 | 1.8 | GO:0031213 | RSF complex(GO:0031213) |
| 0.4 | 3.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 2.9 | GO:1990393 | 3M complex(GO:1990393) |
| 0.4 | 7.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 1.0 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.3 | 1.0 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.2 | 1.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 0.5 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.2 | 1.0 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.7 | GO:0002081 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
| 0.1 | 0.4 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.1 | 0.6 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 1.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 1.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 1.1 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.1 | 0.6 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 1.0 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.1 | 1.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.7 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.2 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 9.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 3.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.1 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.8 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.7 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 1.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 5.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 1.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.0 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 2.5 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 3.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.6 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 7.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.0 | 6.9 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.9 | 3.7 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.6 | 3.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.6 | 1.8 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.4 | 9.1 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.4 | 0.4 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.3 | 8.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 1.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.3 | 5.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 1.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.3 | 2.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 2.0 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 5.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 1.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 2.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 0.7 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 1.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.2 | 1.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.2 | 0.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 2.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 5.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 1.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 1.4 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 3.4 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.4 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 1.0 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 5.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 2.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 1.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.8 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.7 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 3.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.7 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.7 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.3 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 0.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.3 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 2.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.8 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 2.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.5 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.1 | 1.5 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 3.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.0 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.2 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 2.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.1 | 0.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 3.5 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.1 | 0.5 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 0.2 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.5 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 1.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 2.8 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.9 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.1 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 0.1 | 2.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.0 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.7 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 1.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 1.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 1.3 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 1.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.4 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 1.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 3.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.0 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.4 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 5.0 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 5.0 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.1 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID_A6B1_A6B4_INTEGRIN_PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.7 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 7.1 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.8 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.0 | PID_CD8_TCR_DOWNSTREAM_PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.1 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.8 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.5 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 5.7 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 3.1 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 3.4 | REACTOME_XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 5.1 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.1 | REACTOME_INWARDLY_RECTIFYING_K_CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 2.4 | REACTOME_INITIAL_TRIGGERING_OF_COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.0 | REACTOME_ENDOSOMAL_VACUOLAR_PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 2.2 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.7 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 5.8 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.1 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.7 | REACTOME_ABACAVIR_TRANSPORT_AND_METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 3.7 | REACTOME_CDK_MEDIATED_PHOSPHORYLATION_AND_REMOVAL_OF_CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 4.7 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 3.2 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 2.7 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 6.4 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.1 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.1 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.9 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.4 | REACTOME_LIPOPROTEIN_METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.8 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.0 | REACTOME_TRAF6_MEDIATED_NFKB_ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 2.2 | REACTOME_COSTIMULATION_BY_THE_CD28_FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.9 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME_INTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.7 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.5 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME_AUTODEGRADATION_OF_CDH1_BY_CDH1_APC_C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.8 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.2 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME_OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME_BIOSYNTHESIS_OF_THE_N_GLYCAN_PRECURSOR_DOLICHOL_LIPID_LINKED_OLIGOSACCHARIDE_LLO_AND_TRANSFER_TO_A_NASCENT_PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME_THROMBIN_SIGNALLING_THROUGH_PROTEINASE_ACTIVATED_RECEPTORS_PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.3 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.1 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |