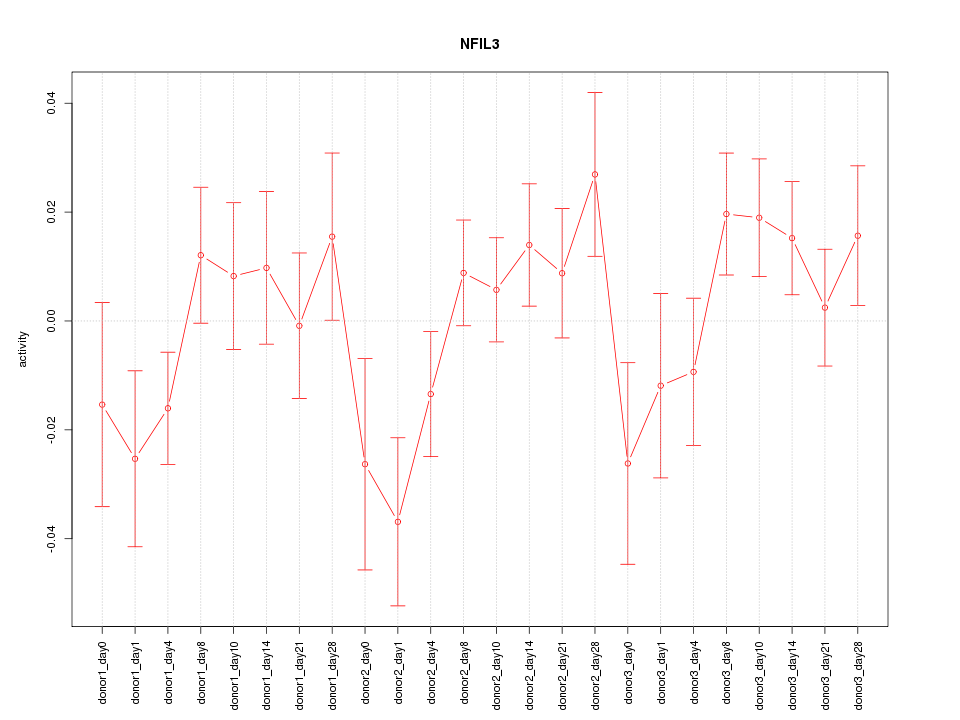
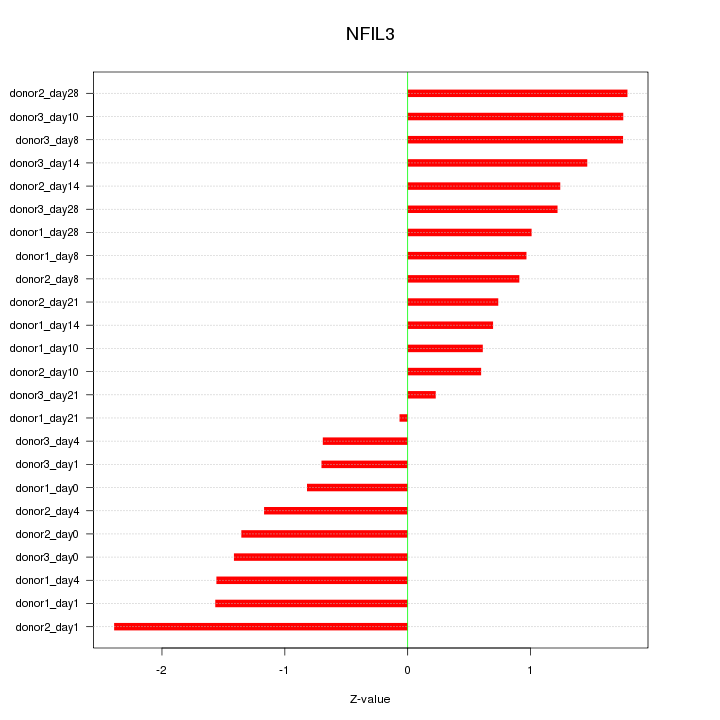
Motif ID: NFIL3
Z-value: 1.234
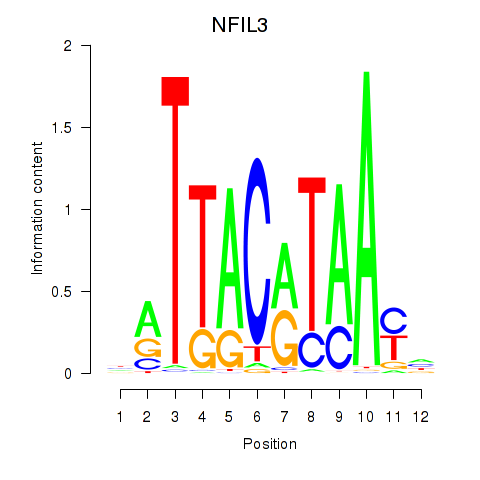
Transcription factors associated with NFIL3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NFIL3 | ENSG00000165030.3 | NFIL3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFIL3 | hg19_v2_chr9_-_94186131_94186174 | -0.55 | 5.7e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 1.2 | 6.0 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 1.1 | 7.4 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.9 | 1.8 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.8 | 2.5 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.7 | 2.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.7 | 3.4 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.7 | 6.0 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.6 | 1.7 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.5 | 1.6 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.5 | 7.6 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.5 | 5.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.5 | 9.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.5 | 1.8 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.4 | 1.7 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.4 | 1.2 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.4 | 4.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.4 | 1.9 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.4 | 3.4 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.4 | 1.1 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.3 | 1.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.3 | 2.0 | GO:1990822 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.3 | 7.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 0.9 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.3 | 1.8 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.3 | 3.3 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.3 | 0.8 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.3 | 1.5 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 0.7 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.2 | 0.5 | GO:0052314 | isoquinoline alkaloid metabolic process(GO:0033076) phytoalexin metabolic process(GO:0052314) |
| 0.2 | 1.4 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.2 | 1.5 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.2 | 0.8 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 1.0 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.2 | 0.4 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.2 | 0.6 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.2 | 1.3 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.2 | 0.6 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.2 | 1.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 3.5 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.2 | 3.0 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.2 | 3.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.2 | 0.8 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.2 | 0.7 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.2 | 0.5 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.2 | 1.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 1.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 0.5 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.2 | 0.5 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.2 | 0.3 | GO:0002537 | nitric oxide production involved in inflammatory response(GO:0002537) |
| 0.2 | 0.6 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 | 2.0 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.4 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 1.6 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.6 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 1.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 1.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.5 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 6.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.8 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.5 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.7 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.1 | 0.2 | GO:0043449 | cellular alkene metabolic process(GO:0043449) olefin metabolic process(GO:1900673) |
| 0.1 | 1.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.5 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.1 | 0.7 | GO:0061052 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 | 2.6 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.1 | 0.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 2.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.4 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 3.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.4 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.5 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.5 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.6 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.1 | 0.9 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 1.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.3 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 1.4 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 4.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.6 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.3 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.5 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 2.0 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 0.3 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.4 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 1.7 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.1 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.3 | GO:0018307 | tRNA wobble position uridine thiolation(GO:0002143) enzyme active site formation(GO:0018307) |
| 0.1 | 0.5 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.6 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 0.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.4 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.3 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 9.8 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 0.4 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 1.1 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.8 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.2 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 0.2 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.1 | 0.2 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.1 | 0.2 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) regulation of SA node cell action potential(GO:0098907) |
| 0.1 | 1.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.9 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.4 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 1.1 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.1 | 0.6 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.1 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.1 | 0.1 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.1 | 1.0 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 0.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.5 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.1 | 0.3 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.1 | 0.7 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 0.2 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.4 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 2.9 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.1 | 0.9 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 1.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.3 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 1.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.7 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 1.0 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.2 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.2 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.1 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.7 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.2 | GO:0051664 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.1 | 1.4 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.7 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.4 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.6 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 1.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.4 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.1 | 0.3 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 1.0 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 1.0 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 1.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.7 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.4 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.4 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.2 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.4 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.2 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0072233 | thick ascending limb development(GO:0072023) distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.5 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.5 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.5 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.4 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 1.1 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.8 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.2 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.2 | GO:1903788 | mycotoxin metabolic process(GO:0043385) mycotoxin catabolic process(GO:0043387) aflatoxin metabolic process(GO:0046222) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound metabolic process(GO:1901376) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 1.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.9 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.4 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 | 0.2 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.0 | 0.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.7 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 2.3 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.7 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.8 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.2 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.0 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 1.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 5.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.4 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 1.3 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:2001027 | negative regulation of lamellipodium assembly(GO:0010593) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.4 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 1.0 | GO:0048863 | stem cell differentiation(GO:0048863) |
| 0.0 | 0.3 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.1 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.0 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.2 | GO:0034670 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.1 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.0 | 0.5 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.4 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 0.4 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.3 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.3 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.2 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 1.1 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.9 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.5 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.8 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.4 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 1.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.5 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 3.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.2 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.4 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 1.0 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.3 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.5 | GO:0090224 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.0 | 1.5 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.6 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.0 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.3 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.1 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.2 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:0035711 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 1.2 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:0015798 | polyol transport(GO:0015791) myo-inositol transport(GO:0015798) |
| 0.0 | 0.4 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.1 | GO:0036499 | PERK-mediated unfolded protein response(GO:0036499) |
| 0.0 | 0.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.4 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 1.0 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.6 | GO:0006505 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.4 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 | 0.2 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.3 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.5 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.4 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.0 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.2 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.3 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.3 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.4 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.8 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.1 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.0 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.3 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.5 | 9.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 1.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 1.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.3 | 1.0 | GO:0030849 | autosome(GO:0030849) |
| 0.3 | 1.9 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.3 | 3.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 10.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 1.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 1.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 2.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 3.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 1.4 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 1.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.3 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.3 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.0 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.6 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.8 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 1.4 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 0.1 | 1.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.5 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 1.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.6 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.7 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.1 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.5 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 4.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 4.7 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 5.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 1.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0044423 | virion(GO:0019012) virion part(GO:0044423) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.0 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 2.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 2.4 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 1.8 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.3 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.5 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.6 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 1.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.4 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.6 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 0.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 15.6 | GO:0042175 | nuclear outer membrane-endoplasmic reticulum membrane network(GO:0042175) |
| 0.0 | 0.0 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 8.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 1.0 | 3.0 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.9 | 3.8 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.7 | 14.7 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.7 | 3.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.6 | 2.5 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.6 | 3.3 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.4 | 1.6 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.4 | 1.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.3 | 1.7 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.3 | 14.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 4.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.3 | 2.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 0.8 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.3 | 4.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 1.6 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.2 | 2.4 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.2 | 1.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 0.9 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.2 | 6.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 1.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.2 | 0.7 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 0.5 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.2 | 1.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.2 | 1.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 0.9 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 0.5 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.7 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.4 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.4 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.1 | 1.2 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.4 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.1 | 2.0 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 1.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.4 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 1.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.8 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.5 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.8 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.3 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.5 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.6 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.6 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.1 | 0.3 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.2 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 3.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 2.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 1.8 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.2 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.1 | 0.7 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.4 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 1.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.7 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.3 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 1.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.1 | 0.8 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.8 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.6 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 0.8 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 3.0 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 0.5 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.1 | 1.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 1.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.5 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.1 | 0.4 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.2 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 3.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 3.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.0 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.3 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.9 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 1.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 1.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 3.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 1.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.9 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 1.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) ion gated channel activity(GO:0022839) |
| 0.0 | 1.0 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.5 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.4 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.4 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.4 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 1.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 1.9 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 1.6 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 1.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.0 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.0 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.6 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID_S1P_S1P2_PATHWAY | S1P2 pathway |
| 0.1 | 1.2 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.1 | 5.6 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.1 | ST_ADRENERGIC | Adrenergic Pathway |
| 0.1 | 0.3 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 4.7 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.8 | PID_TRAIL_PATHWAY | TRAIL signaling pathway |
| 0.1 | 1.9 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.6 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.0 | PID_VEGFR1_PATHWAY | VEGFR1 specific signals |
| 0.0 | 2.2 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.1 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.0 | 0.2 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.7 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.5 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.6 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.4 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.3 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.0 | PID_CD8_TCR_DOWNSTREAM_PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.2 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.2 | PID_FAS_PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 1.1 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.1 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | PID_AR_NONGENOMIC_PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.5 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.3 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | ST_DIFFERENTIATION_PATHWAY_IN_PC12_CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.1 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.0 | 0.6 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 14.8 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.5 | 9.5 | REACTOME_XENOBIOTICS | Genes involved in Xenobiotics |
| 0.3 | 6.0 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 2.3 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 4.8 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.5 | REACTOME_ADP_SIGNALLING_THROUGH_P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 3.5 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 3.0 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.7 | REACTOME_INWARDLY_RECTIFYING_K_CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 1.6 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.0 | REACTOME_OPSINS | Genes involved in Opsins |
| 0.1 | 1.9 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 2.4 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.6 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.1 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.7 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 2.6 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.1 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.0 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.0 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.8 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 2.0 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.3 | REACTOME_ACTIVATION_OF_GENES_BY_ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.0 | REACTOME_DOWNSTREAM_TCR_SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME_THROMBOXANE_SIGNALLING_THROUGH_TP_RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.8 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.8 | REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.9 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.8 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME_THROMBIN_SIGNALLING_THROUGH_PROTEINASE_ACTIVATED_RECEPTORS_PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 1.2 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PLC_BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.5 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.5 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME_REGULATION_OF_KIT_SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.8 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.7 | REACTOME_NEP_NS2_INTERACTS_WITH_THE_CELLULAR_EXPORT_MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.3 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_IFNA_SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.2 | REACTOME_RECYCLING_OF_BILE_ACIDS_AND_SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.9 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.4 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.4 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |