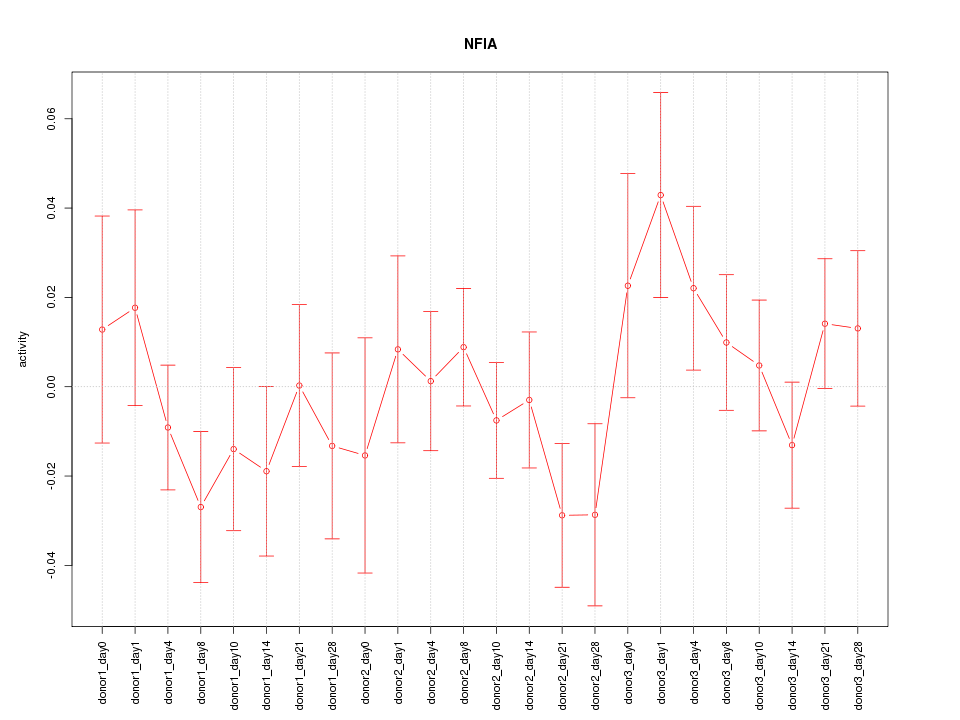
Motif ID: NFIA
Z-value: 0.936
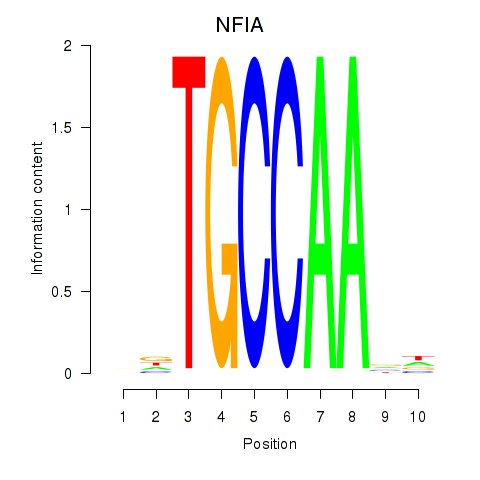
Transcription factors associated with NFIA:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NFIA | ENSG00000162599.11 | NFIA |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFIA | hg19_v2_chr1_+_61869748_61869782 | -0.63 | 9.8e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 1.1 | 4.2 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.7 | 2.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.6 | 2.5 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.6 | 1.8 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.4 | 2.0 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.4 | 1.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.3 | 1.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 | 2.4 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.3 | 0.9 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.3 | 1.7 | GO:2000391 | positive regulation of neutrophil extravasation(GO:2000391) |
| 0.3 | 0.6 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.3 | 1.4 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.3 | 2.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.3 | 1.8 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.2 | 0.7 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.2 | 1.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 0.9 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.2 | 2.4 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.2 | 1.0 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.2 | 0.6 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.2 | 0.6 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.2 | 2.4 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.2 | 1.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 0.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 0.5 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.2 | 1.0 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.2 | 0.8 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.2 | 1.5 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.2 | 4.0 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 1.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 0.4 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.4 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.1 | 0.4 | GO:0060721 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.1 | 0.7 | GO:1904640 | response to methionine(GO:1904640) |
| 0.1 | 0.4 | GO:0098907 | protein localization to M-band(GO:0036309) regulation of SA node cell action potential(GO:0098907) |
| 0.1 | 0.5 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.5 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.4 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 | 1.0 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 0.4 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.5 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) |
| 0.1 | 0.6 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.6 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.6 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 0.3 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.6 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.1 | 0.3 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 0.6 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.4 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 0.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 1.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 0.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.5 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.1 | 0.3 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.4 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.9 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.3 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.6 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.8 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 1.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.2 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.3 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.1 | 0.2 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.1 | 0.3 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.2 | GO:0090294 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.1 | 0.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.3 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.1 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 4.7 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 0.2 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.5 | GO:2000782 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) regulation of barbed-end actin filament capping(GO:2000812) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.1 | 0.9 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 0.3 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.1 | 0.4 | GO:0033504 | floor plate development(GO:0033504) |
| 0.1 | 1.3 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.4 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.1 | 0.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.2 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 0.6 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.1 | 0.2 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 0.8 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 2.9 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.3 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 1.8 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 0.4 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.5 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.0 | 0.3 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.4 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.2 | GO:0097021 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.1 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.3 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.2 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.5 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.7 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.4 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.2 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.4 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 1.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.4 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.6 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.0 | 0.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 1.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) glycerol biosynthetic process(GO:0006114) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.3 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.8 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0046440 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.7 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.6 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0042746 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.0 | 0.7 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 1.7 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.5 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 2.9 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.8 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.6 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.2 | GO:0031329 | regulation of cellular catabolic process(GO:0031329) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.2 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.4 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0071865 | regulation of apoptotic process in bone marrow(GO:0071865) negative regulation of apoptotic process in bone marrow(GO:0071866) |
| 0.0 | 0.3 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.5 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.9 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.2 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.4 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 1.9 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.5 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 4.9 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.0 | 1.6 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.5 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.4 | GO:0050673 | epithelial cell proliferation(GO:0050673) |
| 0.0 | 0.0 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.0 | GO:1900110 | regulation of histone H3-K9 dimethylation(GO:1900109) negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 | 0.1 | GO:0071415 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.0 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.1 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.2 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.5 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.4 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.0 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.5 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 1.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 1.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.3 | 2.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.2 | 2.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.1 | 2.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 2.0 | GO:0033643 | host cell part(GO:0033643) |
| 0.1 | 0.3 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 0.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.3 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.1 | 5.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.6 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 2.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.5 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 1.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 2.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 2.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 2.1 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.0 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 2.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 1.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.7 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.5 | 2.5 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.5 | 4.2 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.4 | 2.0 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.4 | 1.6 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.4 | 1.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 1.8 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.3 | 1.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.3 | 1.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.3 | 1.8 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.3 | 0.9 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.2 | 2.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 0.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 1.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 0.5 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.2 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.4 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 2.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.4 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 0.4 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.5 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.5 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 1.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.1 | 0.4 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.9 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.7 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.5 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 1.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 2.0 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 0.2 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.1 | 1.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 2.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.6 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 1.0 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.2 | GO:0019961 | interferon binding(GO:0019961) |
| 0.1 | 0.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.2 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 1.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.3 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 2.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.6 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.5 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.9 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) phosphatidylserine decarboxylase activity(GO:0004609) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 1.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.6 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 3.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.2 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 2.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.6 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 2.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 3.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 1.2 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.0 | 0.0 | GO:0031493 | nucleosomal histone binding(GO:0031493) SAM domain binding(GO:0032093) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 2.3 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.9 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.1 | 2.0 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.5 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 4.5 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.4 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.9 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.8 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.9 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.3 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.5 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | PID_IL2_1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.8 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.4 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | ST_TYPE_I_INTERFERON_PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.5 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.6 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 1.7 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID_EPO_PATHWAY | EPO signaling pathway |
| 0.0 | 1.2 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 3.6 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.6 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.8 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 4.2 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 2.2 | REACTOME_ANDROGEN_BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 2.8 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.4 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.8 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.6 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.7 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_24_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME_TRAFFICKING_AND_PROCESSING_OF_ENDOSOMAL_TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.7 | REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.9 | REACTOME_STEROID_HORMONES | Genes involved in Steroid hormones |
| 0.0 | 2.2 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.7 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.6 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.8 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.0 | 1.4 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.6 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.7 | REACTOME_SEMA3A_PAK_DEPENDENT_AXON_REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.3 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.9 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.3 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.4 | REACTOME_TRYPTOPHAN_CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME_METABOLISM_OF_STEROID_HORMONES_AND_VITAMINS_A_AND_D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME_ENDOSOMAL_VACUOLAR_PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME_UNBLOCKING_OF_NMDA_RECEPTOR_GLUTAMATE_BINDING_AND_ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 1.3 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |