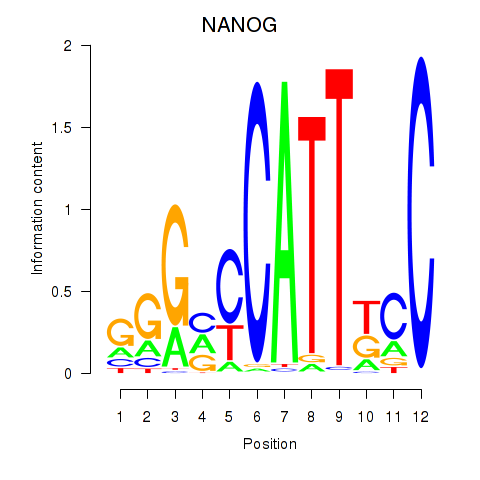
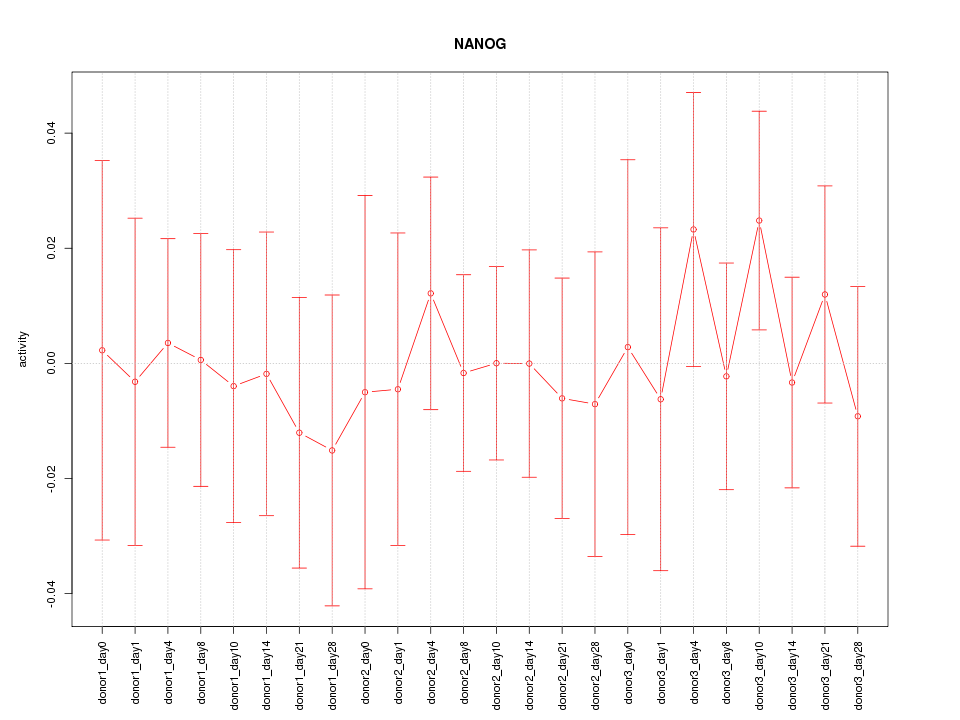
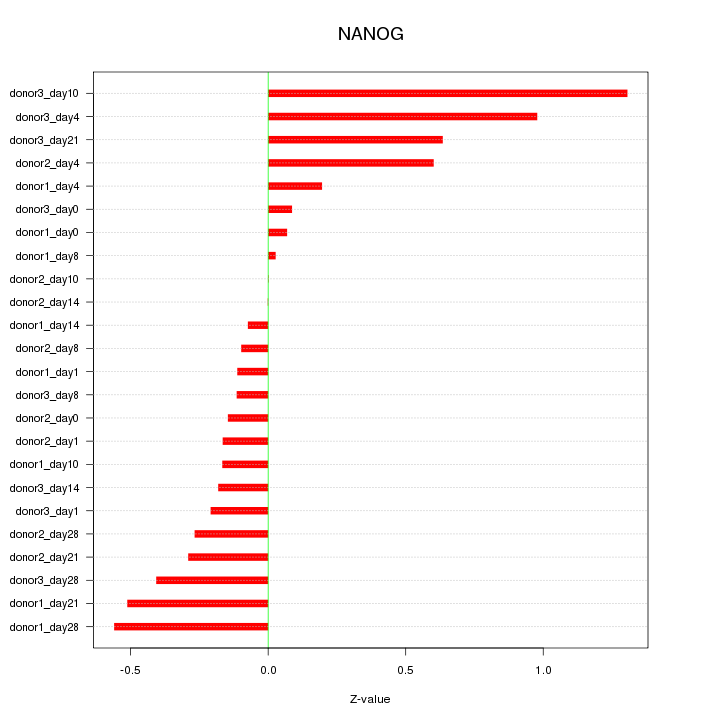
| 0.1 |
0.5 |
GO:0033031 |
positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 |
0.2 |
GO:1904298 |
positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 0.1 |
0.2 |
GO:0070681 |
glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 |
0.1 |
GO:0032242 |
regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) |
| 0.0 |
0.1 |
GO:1904617 |
negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 |
0.1 |
GO:0070253 |
somatostatin secretion(GO:0070253) |
| 0.0 |
0.1 |
GO:0002644 |
negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 |
0.2 |
GO:0032417 |
positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 |
0.1 |
GO:0031296 |
positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 |
0.8 |
GO:0034380 |
cartilage condensation(GO:0001502) high-density lipoprotein particle assembly(GO:0034380) |
| 0.0 |
0.2 |
GO:0035502 |
metanephric part of ureteric bud development(GO:0035502) |
| 0.0 |
0.2 |
GO:0070236 |
negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 |
0.1 |
GO:0001172 |
transcription, RNA-templated(GO:0001172) |
| 0.0 |
0.1 |
GO:0090402 |
oncogene-induced cell senescence(GO:0090402) |
| 0.0 |
0.1 |
GO:0018352 |
protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 |
0.2 |
GO:2000543 |
positive regulation of gastrulation(GO:2000543) |
| 0.0 |
0.1 |
GO:2000503 |
positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 |
0.1 |
GO:0018879 |
biphenyl metabolic process(GO:0018879) |
| 0.0 |
0.1 |
GO:0042732 |
D-xylose metabolic process(GO:0042732) |
| 0.0 |
0.1 |
GO:1903989 |
regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 |
0.0 |
GO:0002316 |
follicular B cell differentiation(GO:0002316) |
| 0.0 |
0.0 |
GO:2000144 |
positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 |
0.1 |
GO:0009082 |
branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 |
0.1 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 |
0.3 |
GO:0045141 |
meiotic telomere clustering(GO:0045141) |
| 0.0 |
0.6 |
GO:0032958 |
inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 |
0.2 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 |
0.1 |
GO:0033211 |
adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 |
0.0 |
GO:1904717 |
excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 |
0.1 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |