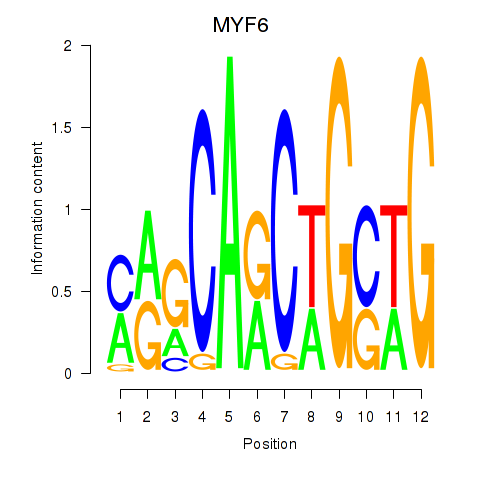
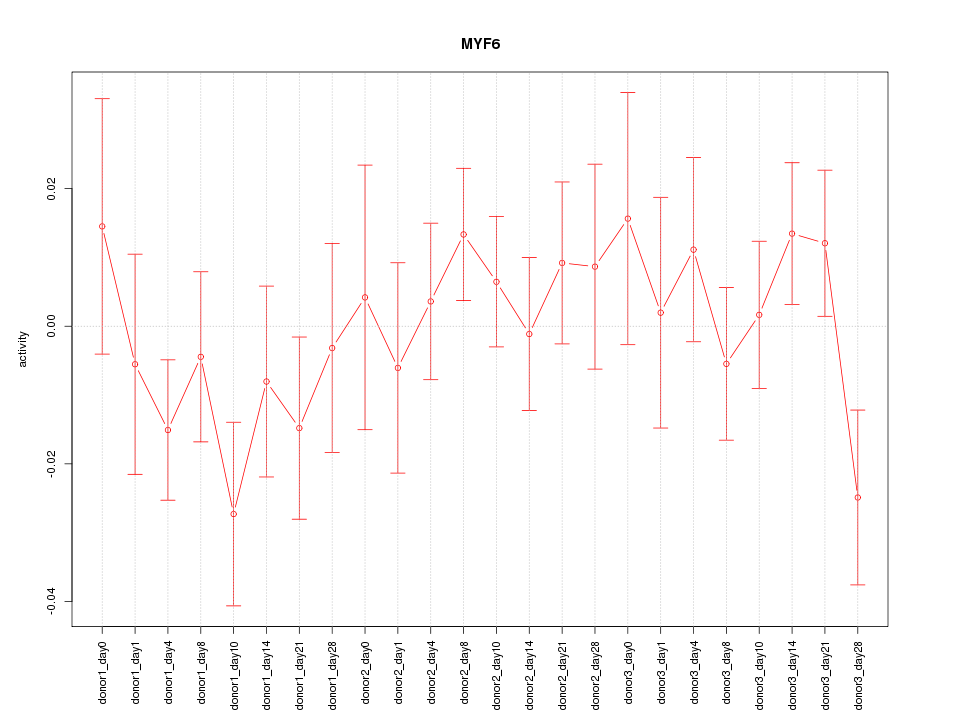
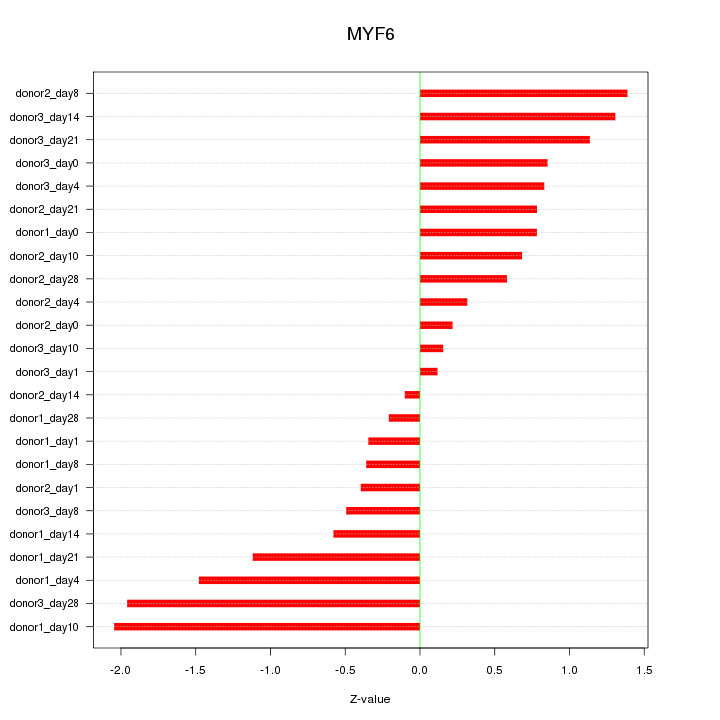
| 0.9 |
2.8 |
GO:0042369 |
vitamin D catabolic process(GO:0042369) |
| 0.4 |
1.2 |
GO:0090271 |
positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.4 |
3.1 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.4 |
1.4 |
GO:1902460 |
regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.3 |
1.0 |
GO:0045210 |
FasL biosynthetic process(GO:0045210) |
| 0.3 |
0.9 |
GO:0060435 |
bronchiole development(GO:0060435) |
| 0.3 |
0.9 |
GO:0071109 |
superior temporal gyrus development(GO:0071109) |
| 0.3 |
0.9 |
GO:0015920 |
lipopolysaccharide transport(GO:0015920) |
| 0.3 |
0.8 |
GO:0052047 |
interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.2 |
0.4 |
GO:0097252 |
oligodendrocyte apoptotic process(GO:0097252) |
| 0.2 |
0.7 |
GO:0021966 |
corticospinal neuron axon guidance(GO:0021966) |
| 0.2 |
0.6 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.2 |
1.7 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 |
0.6 |
GO:1904397 |
negative regulation of neuromuscular junction development(GO:1904397) |
| 0.2 |
0.6 |
GO:0046947 |
hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 |
0.5 |
GO:0017186 |
peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 |
0.4 |
GO:2000646 |
positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 |
0.6 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 |
0.5 |
GO:0072268 |
pattern specification involved in metanephros development(GO:0072268) |
| 0.1 |
0.4 |
GO:0019072 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 |
0.4 |
GO:0090341 |
negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.1 |
0.4 |
GO:0043397 |
corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 |
0.7 |
GO:0055064 |
chloride ion homeostasis(GO:0055064) |
| 0.1 |
0.3 |
GO:0042700 |
luteinizing hormone signaling pathway(GO:0042700) |
| 0.1 |
0.6 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.1 |
0.4 |
GO:0036378 |
calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.1 |
0.5 |
GO:1901662 |
phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 |
0.6 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 |
0.3 |
GO:0032824 |
natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 |
0.6 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
0.4 |
GO:0045065 |
cytotoxic T cell differentiation(GO:0045065) |
| 0.1 |
0.8 |
GO:0070236 |
negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 |
0.4 |
GO:0038195 |
urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 |
0.3 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.1 |
0.3 |
GO:0042264 |
peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 |
0.4 |
GO:0061143 |
alveolar primary septum development(GO:0061143) |
| 0.1 |
0.2 |
GO:0055073 |
cadmium ion homeostasis(GO:0055073) lysosomal membrane organization(GO:0097212) negative regulation of hydrogen peroxide catabolic process(GO:2000296) regulation of oxygen metabolic process(GO:2000374) |
| 0.1 |
0.7 |
GO:0060754 |
positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 |
1.0 |
GO:0060083 |
smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 |
0.1 |
GO:1903862 |
positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 |
0.2 |
GO:0046521 |
sphingoid catabolic process(GO:0046521) |
| 0.1 |
0.3 |
GO:1905167 |
positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 |
0.2 |
GO:1904339 |
negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 |
0.4 |
GO:0021773 |
striatal medium spiny neuron differentiation(GO:0021773) |
| 0.1 |
0.5 |
GO:0021842 |
directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 |
0.2 |
GO:0070235 |
CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) regulation of activation-induced cell death of T cells(GO:0070235) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.1 |
0.2 |
GO:0048015 |
phosphatidylinositol-mediated signaling(GO:0048015) inositol lipid-mediated signaling(GO:0048017) |
| 0.1 |
0.3 |
GO:0000711 |
meiotic DNA repair synthesis(GO:0000711) |
| 0.1 |
0.2 |
GO:0090187 |
positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 |
0.2 |
GO:0021849 |
neuroblast division in subventricular zone(GO:0021849) |
| 0.1 |
0.2 |
GO:0035493 |
SNARE complex assembly(GO:0035493) |
| 0.1 |
0.4 |
GO:0035407 |
histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 |
0.3 |
GO:0002378 |
immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 |
0.6 |
GO:2000252 |
negative regulation of feeding behavior(GO:2000252) |
| 0.1 |
0.4 |
GO:0060125 |
negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 |
0.2 |
GO:0031052 |
mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.1 |
0.2 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 |
0.2 |
GO:0014878 |
response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 |
0.7 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.1 |
0.3 |
GO:1902766 |
skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 |
1.0 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 |
0.3 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 |
0.3 |
GO:0072233 |
thick ascending limb development(GO:0072023) DCT cell differentiation(GO:0072069) metanephric thick ascending limb development(GO:0072233) metanephric DCT cell differentiation(GO:0072240) |
| 0.1 |
0.4 |
GO:2001137 |
positive regulation of endocytic recycling(GO:2001137) |
| 0.1 |
10.7 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.1 |
0.3 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.1 |
0.2 |
GO:0021526 |
medial motor column neuron differentiation(GO:0021526) |
| 0.1 |
0.2 |
GO:1905123 |
regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.1 |
0.3 |
GO:0033274 |
response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 |
0.2 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.1 |
0.2 |
GO:0021987 |
cerebral cortex development(GO:0021987) |
| 0.0 |
0.1 |
GO:1903697 |
negative regulation of microvillus assembly(GO:1903697) |
| 0.0 |
0.2 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 |
0.2 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.0 |
0.3 |
GO:0098704 |
fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 |
0.2 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.1 |
GO:0046166 |
glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 |
0.1 |
GO:2000182 |
regulation of progesterone biosynthetic process(GO:2000182) |
| 0.0 |
0.2 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 |
0.2 |
GO:0090031 |
positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 |
0.1 |
GO:0043449 |
olfactory learning(GO:0008355) cellular alkene metabolic process(GO:0043449) |
| 0.0 |
0.2 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 |
0.2 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 |
0.2 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.0 |
0.3 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) |
| 0.0 |
0.0 |
GO:0015680 |
intracellular copper ion transport(GO:0015680) |
| 0.0 |
0.1 |
GO:0042701 |
progesterone secretion(GO:0042701) |
| 0.0 |
0.4 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.2 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 |
0.3 |
GO:0032224 |
positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 |
0.1 |
GO:0051835 |
positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 |
0.1 |
GO:0016340 |
calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 |
0.2 |
GO:1902963 |
regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 |
0.3 |
GO:0043932 |
ossification involved in bone remodeling(GO:0043932) |
| 0.0 |
0.1 |
GO:0038193 |
thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 |
0.0 |
GO:0021539 |
subthalamus development(GO:0021539) |
| 0.0 |
0.1 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 |
0.1 |
GO:0060265 |
positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.0 |
0.4 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.0 |
0.1 |
GO:0044209 |
AMP salvage(GO:0044209) |
| 0.0 |
0.1 |
GO:1903989 |
regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 |
0.4 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.0 |
0.1 |
GO:0001880 |
Mullerian duct regression(GO:0001880) regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 |
0.1 |
GO:0046900 |
tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 |
0.2 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 |
0.2 |
GO:0070309 |
lens fiber cell morphogenesis(GO:0070309) |
| 0.0 |
0.2 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 |
0.2 |
GO:2000382 |
positive regulation of mesoderm development(GO:2000382) |
| 0.0 |
0.1 |
GO:0048739 |
cardiac muscle fiber development(GO:0048739) |
| 0.0 |
0.4 |
GO:0008212 |
mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 |
0.1 |
GO:1990167 |
protein K27-linked deubiquitination(GO:1990167) |
| 0.0 |
0.1 |
GO:1900369 |
transcription, RNA-templated(GO:0001172) negative regulation of RNA interference(GO:1900369) |
| 0.0 |
0.2 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.0 |
0.2 |
GO:0006537 |
glutamate biosynthetic process(GO:0006537) |
| 0.0 |
0.2 |
GO:0035754 |
B cell chemotaxis(GO:0035754) |
| 0.0 |
0.1 |
GO:0018242 |
protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 |
0.2 |
GO:0097084 |
vascular smooth muscle cell development(GO:0097084) |
| 0.0 |
0.1 |
GO:0032455 |
nerve growth factor processing(GO:0032455) |
| 0.0 |
0.1 |
GO:1903377 |
negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 |
0.5 |
GO:0010623 |
programmed cell death involved in cell development(GO:0010623) |
| 0.0 |
0.4 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 |
0.1 |
GO:1903566 |
positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 |
0.1 |
GO:0032425 |
positive regulation of mismatch repair(GO:0032425) |
| 0.0 |
0.1 |
GO:0016476 |
regulation of embryonic cell shape(GO:0016476) |
| 0.0 |
0.1 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 |
0.1 |
GO:0002277 |
myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.0 |
0.2 |
GO:1905247 |
positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 |
0.6 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 |
0.3 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.0 |
0.1 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.0 |
0.1 |
GO:0019470 |
4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 |
0.3 |
GO:2001013 |
epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 |
0.5 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.0 |
0.1 |
GO:0002879 |
positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 |
0.1 |
GO:0035690 |
cellular response to drug(GO:0035690) |
| 0.0 |
0.6 |
GO:0034755 |
iron ion transmembrane transport(GO:0034755) |
| 0.0 |
0.2 |
GO:1904715 |
negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 |
0.1 |
GO:0097411 |
hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 |
0.2 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.1 |
GO:0061056 |
sclerotome development(GO:0061056) |
| 0.0 |
0.3 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.0 |
0.4 |
GO:0006878 |
cellular copper ion homeostasis(GO:0006878) |
| 0.0 |
0.1 |
GO:0060586 |
multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 |
0.1 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.0 |
0.2 |
GO:0070574 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 |
0.3 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.0 |
0.3 |
GO:0031580 |
membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 |
0.6 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.0 |
GO:0009609 |
response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.0 |
0.1 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.0 |
0.1 |
GO:0048842 |
positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 |
0.1 |
GO:0003358 |
noradrenergic neuron development(GO:0003358) |
| 0.0 |
0.2 |
GO:1905146 |
lysosomal protein catabolic process(GO:1905146) |
| 0.0 |
0.1 |
GO:0038169 |
somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 |
0.3 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.0 |
0.3 |
GO:0019371 |
cyclooxygenase pathway(GO:0019371) |
| 0.0 |
0.1 |
GO:0036269 |
swimming behavior(GO:0036269) |
| 0.0 |
0.2 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.1 |
GO:0060988 |
lipid tube assembly(GO:0060988) |
| 0.0 |
0.1 |
GO:0061762 |
CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 |
0.3 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 |
0.1 |
GO:0003222 |
ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.0 |
0.1 |
GO:0032185 |
septin cytoskeleton organization(GO:0032185) |
| 0.0 |
0.1 |
GO:0033227 |
dsRNA transport(GO:0033227) |
| 0.0 |
0.0 |
GO:2000566 |
positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 |
0.1 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 |
0.1 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 |
0.1 |
GO:0060137 |
maternal process involved in parturition(GO:0060137) |
| 0.0 |
0.1 |
GO:0034436 |
glycoprotein transport(GO:0034436) |
| 0.0 |
0.1 |
GO:2000568 |
memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 |
0.1 |
GO:0048631 |
regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 |
0.1 |
GO:0032808 |
lacrimal gland development(GO:0032808) |
| 0.0 |
0.2 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.1 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.8 |
GO:0090004 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 |
0.2 |
GO:0045663 |
positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 |
0.0 |
GO:0090427 |
activation of meiosis(GO:0090427) |
| 0.0 |
0.1 |
GO:2001206 |
positive regulation of osteoclast development(GO:2001206) |
| 0.0 |
0.2 |
GO:0060897 |
neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 |
0.2 |
GO:0003344 |
pericardium morphogenesis(GO:0003344) |
| 0.0 |
0.1 |
GO:0071321 |
cellular response to cGMP(GO:0071321) |
| 0.0 |
0.4 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 |
0.1 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 |
0.1 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.0 |
0.0 |
GO:1902995 |
regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.0 |
0.1 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.1 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.0 |
GO:0050894 |
determination of affect(GO:0050894) |
| 0.0 |
0.6 |
GO:0010831 |
positive regulation of myotube differentiation(GO:0010831) |
| 0.0 |
0.1 |
GO:0036496 |
regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 |
0.1 |
GO:0070561 |
vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 |
0.1 |
GO:1903423 |
positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle transport(GO:1902805) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 |
0.1 |
GO:0042116 |
macrophage activation(GO:0042116) |
| 0.0 |
0.2 |
GO:0000186 |
activation of MAPKK activity(GO:0000186) |
| 0.0 |
0.1 |
GO:0050779 |
RNA destabilization(GO:0050779) |
| 0.0 |
0.2 |
GO:0045657 |
positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 |
0.2 |
GO:0003229 |
ventricular cardiac muscle tissue development(GO:0003229) |
| 0.0 |
0.1 |
GO:1904783 |
positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.0 |
0.0 |
GO:1902474 |
positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 |
0.1 |
GO:0050916 |
sensory perception of sweet taste(GO:0050916) |
| 0.0 |
0.1 |
GO:2000304 |
positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 |
0.2 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.0 |
0.0 |
GO:0019056 |
modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 |
0.1 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.4 |
GO:0060445 |
branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 |
0.1 |
GO:0031998 |
regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 |
0.3 |
GO:0032933 |
response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 |
0.1 |
GO:0050668 |
cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 |
0.2 |
GO:0036066 |
protein O-linked fucosylation(GO:0036066) |
| 0.0 |
0.1 |
GO:0043116 |
negative regulation of vascular permeability(GO:0043116) |
| 0.0 |
0.1 |
GO:0051149 |
positive regulation of muscle cell differentiation(GO:0051149) |
| 0.0 |
0.1 |
GO:0050717 |
positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 |
0.1 |
GO:0050849 |
negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 |
0.8 |
GO:0045540 |
regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 |
0.0 |
GO:0002339 |
positive regulation of type I hypersensitivity(GO:0001812) B cell selection(GO:0002339) |
| 0.0 |
0.1 |
GO:0050893 |
sensory processing(GO:0050893) |
| 0.0 |
0.4 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.1 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.0 |
0.0 |
GO:0071314 |
cellular response to cocaine(GO:0071314) |
| 0.0 |
0.3 |
GO:0070262 |
peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 |
0.1 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
0.1 |
GO:0003096 |
renal sodium ion transport(GO:0003096) |
| 0.0 |
0.5 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 |
0.2 |
GO:0043517 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 |
0.3 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 |
0.1 |
GO:0000432 |
regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 |
0.2 |
GO:0003085 |
negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.0 |
0.3 |
GO:0030252 |
growth hormone secretion(GO:0030252) |
| 0.0 |
0.5 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.0 |
GO:0006408 |
snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 |
0.1 |
GO:0006642 |
triglyceride mobilization(GO:0006642) |
| 0.0 |
0.5 |
GO:0051693 |
actin filament capping(GO:0051693) |
| 0.0 |
0.0 |
GO:0007181 |
ventricular compact myocardium morphogenesis(GO:0003223) transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 |
0.2 |
GO:0098703 |
calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 |
0.1 |
GO:2000553 |
positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.0 |
0.2 |
GO:0042340 |
keratan sulfate catabolic process(GO:0042340) |
| 0.0 |
0.0 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.0 |
0.1 |
GO:0048304 |
positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 |
0.2 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.0 |
0.0 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 |
0.1 |
GO:2000738 |
positive regulation of stem cell differentiation(GO:2000738) |
| 0.0 |
0.4 |
GO:0050919 |
negative chemotaxis(GO:0050919) |
| 0.0 |
0.4 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.0 |
0.2 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 |
0.1 |
GO:0060259 |
regulation of feeding behavior(GO:0060259) |
| 0.0 |
0.2 |
GO:1903830 |
magnesium ion transmembrane transport(GO:1903830) |
| 0.0 |
0.0 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 |
0.1 |
GO:0045956 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 |
0.2 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |