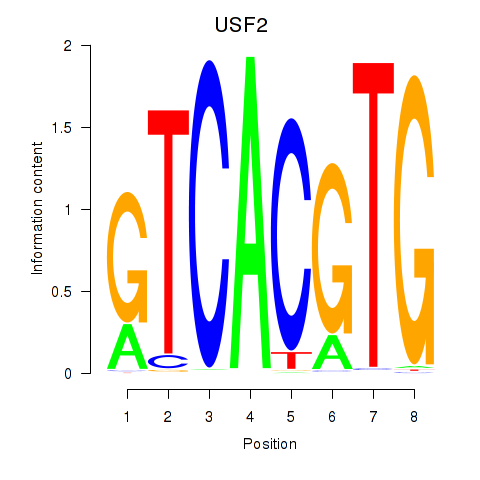
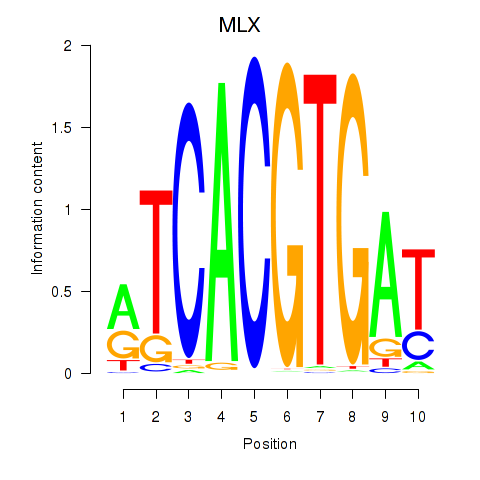
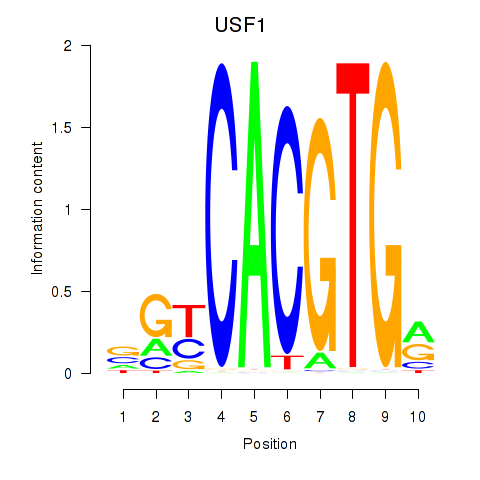
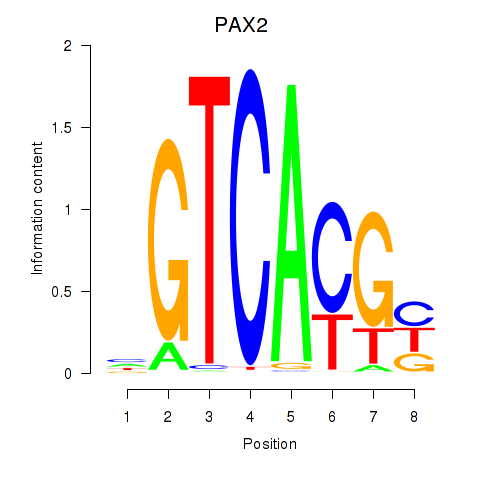
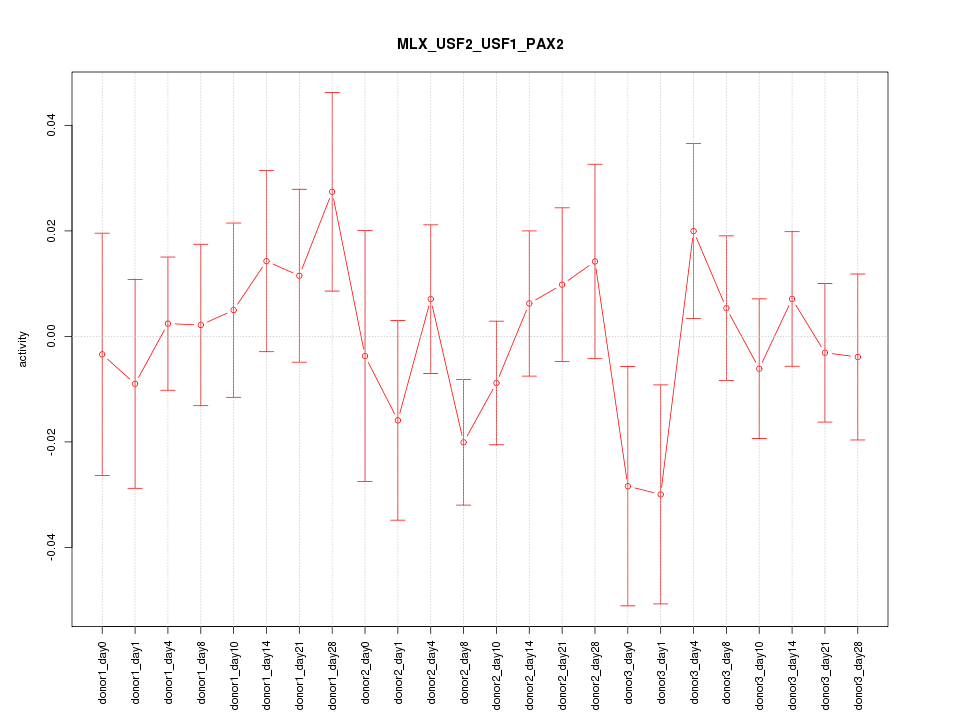
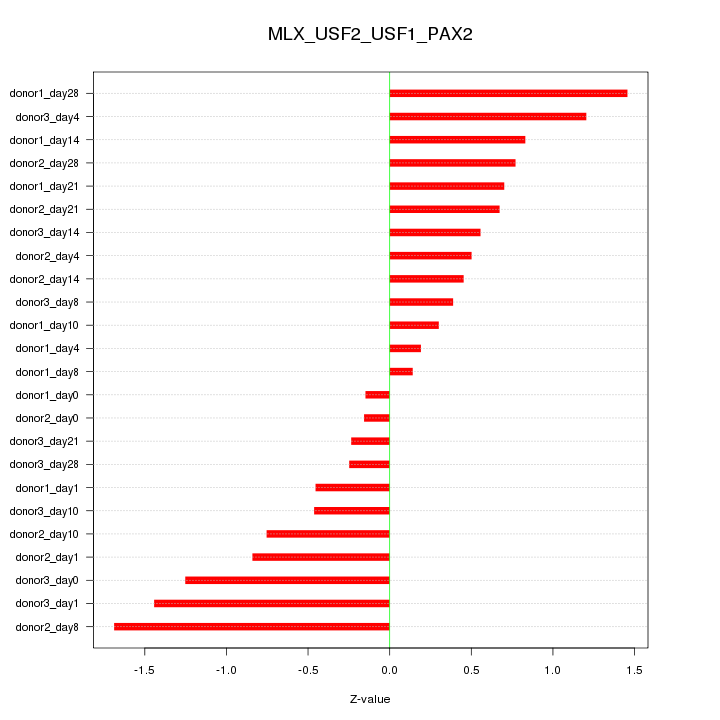
| 1.2 |
6.2 |
GO:0002415 |
immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.7 |
2.0 |
GO:1900169 |
regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.6 |
1.8 |
GO:0006864 |
pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.6 |
3.0 |
GO:0015670 |
carbon dioxide transport(GO:0015670) |
| 0.6 |
5.3 |
GO:1990253 |
cellular response to leucine starvation(GO:1990253) |
| 0.6 |
1.7 |
GO:1905072 |
apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.5 |
1.5 |
GO:0006059 |
hexitol metabolic process(GO:0006059) |
| 0.5 |
1.5 |
GO:0051097 |
negative regulation of helicase activity(GO:0051097) |
| 0.5 |
1.5 |
GO:0035604 |
fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.5 |
9.6 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.4 |
1.3 |
GO:1903568 |
negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.4 |
1.6 |
GO:0060830 |
ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.4 |
3.9 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.4 |
1.1 |
GO:0006788 |
heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.4 |
1.8 |
GO:0006740 |
NADPH regeneration(GO:0006740) |
| 0.3 |
1.6 |
GO:0045629 |
negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.3 |
1.2 |
GO:0006210 |
pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.3 |
1.2 |
GO:0010157 |
response to chlorate(GO:0010157) |
| 0.2 |
1.7 |
GO:0097338 |
response to clozapine(GO:0097338) |
| 0.2 |
0.7 |
GO:0035494 |
SNARE complex disassembly(GO:0035494) |
| 0.2 |
0.7 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 |
0.6 |
GO:0010845 |
positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.2 |
0.6 |
GO:0060086 |
circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.2 |
0.6 |
GO:1901860 |
positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.2 |
1.5 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 |
0.8 |
GO:0032053 |
ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 |
0.6 |
GO:0016340 |
calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.2 |
0.6 |
GO:0070428 |
regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.2 |
0.6 |
GO:0000472 |
endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.2 |
0.7 |
GO:0005986 |
sucrose biosynthetic process(GO:0005986) |
| 0.2 |
0.5 |
GO:0071284 |
cellular response to lead ion(GO:0071284) |
| 0.2 |
1.0 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.2 |
0.5 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.2 |
0.5 |
GO:1902869 |
commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) regulation of amacrine cell differentiation(GO:1902869) |
| 0.2 |
2.5 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.2 |
0.5 |
GO:1904933 |
regulation of cell proliferation in midbrain(GO:1904933) |
| 0.2 |
0.3 |
GO:0002572 |
pro-T cell differentiation(GO:0002572) |
| 0.2 |
0.5 |
GO:0036353 |
histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 |
0.8 |
GO:2000619 |
negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.1 |
0.7 |
GO:0032687 |
negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 |
0.7 |
GO:1900113 |
negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 |
0.8 |
GO:0035407 |
histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 |
0.9 |
GO:0033183 |
negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 |
1.4 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.1 |
1.0 |
GO:0050893 |
sensory processing(GO:0050893) |
| 0.1 |
3.1 |
GO:0032703 |
negative regulation of interleukin-2 production(GO:0032703) |
| 0.1 |
0.7 |
GO:0061484 |
hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 |
0.9 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) |
| 0.1 |
0.2 |
GO:0099552 |
trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 |
0.3 |
GO:2001245 |
regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 |
0.4 |
GO:0000255 |
allantoin metabolic process(GO:0000255) |
| 0.1 |
0.3 |
GO:0072095 |
regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.1 |
0.3 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 |
1.3 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 |
0.2 |
GO:1903348 |
positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 |
0.4 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 |
1.6 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.1 |
1.6 |
GO:0046185 |
aldehyde catabolic process(GO:0046185) |
| 0.1 |
0.4 |
GO:0000957 |
mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 |
0.4 |
GO:0048041 |
cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.1 |
1.2 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.1 |
0.3 |
GO:1905246 |
regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 |
0.4 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 |
0.4 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 |
0.3 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 |
0.3 |
GO:1904327 |
protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 |
0.1 |
GO:0015846 |
polyamine transport(GO:0015846) |
| 0.1 |
0.3 |
GO:0044837 |
assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 |
1.2 |
GO:0032688 |
negative regulation of interferon-beta production(GO:0032688) |
| 0.1 |
0.6 |
GO:0098535 |
de novo centriole assembly(GO:0098535) |
| 0.1 |
0.6 |
GO:0051106 |
positive regulation of DNA ligation(GO:0051106) |
| 0.1 |
0.2 |
GO:0061055 |
myotome development(GO:0061055) |
| 0.1 |
1.3 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.1 |
0.5 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.1 |
0.3 |
GO:0001970 |
positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.1 |
0.3 |
GO:0030037 |
actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 |
1.4 |
GO:0042340 |
keratan sulfate catabolic process(GO:0042340) |
| 0.1 |
1.2 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 |
0.9 |
GO:0021520 |
spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 |
0.5 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 |
0.2 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 |
0.2 |
GO:0046167 |
glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 |
0.2 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.1 |
0.2 |
GO:1904760 |
myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.1 |
0.7 |
GO:0019511 |
peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 |
0.3 |
GO:0046462 |
monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 |
0.2 |
GO:0034059 |
response to anoxia(GO:0034059) |
| 0.1 |
0.3 |
GO:0043335 |
protein unfolding(GO:0043335) |
| 0.1 |
1.7 |
GO:0016254 |
preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 |
0.3 |
GO:0043324 |
eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.1 |
0.3 |
GO:0010748 |
regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 |
0.5 |
GO:0034644 |
cellular response to UV(GO:0034644) |
| 0.1 |
0.2 |
GO:0002752 |
cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 |
0.5 |
GO:1904491 |
protein localization to ciliary transition zone(GO:1904491) |
| 0.1 |
0.3 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.1 |
0.2 |
GO:1900073 |
regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.1 |
0.2 |
GO:0006286 |
base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.1 |
0.5 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.1 |
0.3 |
GO:0008588 |
release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 |
0.1 |
GO:0044828 |
negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 |
0.4 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.1 |
0.7 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 |
1.4 |
GO:0090051 |
negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.1 |
0.2 |
GO:0060748 |
tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.1 |
0.4 |
GO:0006651 |
diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 |
0.1 |
GO:0061289 |
cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 |
0.6 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 |
0.6 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 |
0.3 |
GO:0045345 |
positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.1 |
0.2 |
GO:0048698 |
negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 |
0.3 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 |
0.3 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 |
0.5 |
GO:0046465 |
dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.1 |
0.2 |
GO:0018057 |
peptidyl-lysine oxidation(GO:0018057) |
| 0.1 |
0.2 |
GO:0021571 |
rhombomere 5 development(GO:0021571) |
| 0.1 |
0.1 |
GO:0005985 |
sucrose metabolic process(GO:0005985) |
| 0.1 |
0.2 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 |
0.7 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 |
0.4 |
GO:1990034 |
cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 |
0.5 |
GO:1903800 |
positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 |
0.7 |
GO:0051256 |
mitotic spindle midzone assembly(GO:0051256) |
| 0.1 |
1.7 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.1 |
0.2 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 |
1.1 |
GO:1900745 |
positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 |
0.1 |
GO:1901069 |
guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 |
0.5 |
GO:0001714 |
endodermal cell fate specification(GO:0001714) |
| 0.0 |
0.2 |
GO:0032264 |
IMP salvage(GO:0032264) |
| 0.0 |
0.3 |
GO:0043587 |
tongue morphogenesis(GO:0043587) |
| 0.0 |
0.1 |
GO:0070105 |
positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 |
2.7 |
GO:1904031 |
positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 |
0.2 |
GO:0044565 |
dendritic cell proliferation(GO:0044565) |
| 0.0 |
0.1 |
GO:0072720 |
response to dithiothreitol(GO:0072720) |
| 0.0 |
0.2 |
GO:1902044 |
regulation of Fas signaling pathway(GO:1902044) |
| 0.0 |
0.9 |
GO:0030575 |
nuclear body organization(GO:0030575) |
| 0.0 |
0.2 |
GO:1904715 |
negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 |
1.6 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.0 |
0.2 |
GO:0072719 |
cellular response to cisplatin(GO:0072719) |
| 0.0 |
0.3 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 |
0.3 |
GO:0034128 |
negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 |
0.1 |
GO:0009268 |
response to pH(GO:0009268) |
| 0.0 |
0.2 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.0 |
0.5 |
GO:0034058 |
endosomal vesicle fusion(GO:0034058) |
| 0.0 |
1.1 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
0.7 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 |
0.2 |
GO:0009386 |
translational attenuation(GO:0009386) |
| 0.0 |
0.3 |
GO:0009838 |
abscission(GO:0009838) |
| 0.0 |
0.2 |
GO:0032474 |
otolith morphogenesis(GO:0032474) |
| 0.0 |
0.1 |
GO:0010936 |
negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 |
0.2 |
GO:0010730 |
negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 |
0.2 |
GO:0061209 |
cell proliferation involved in mesonephros development(GO:0061209) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 0.0 |
0.0 |
GO:0090283 |
regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 |
0.2 |
GO:0031296 |
B cell costimulation(GO:0031296) |
| 0.0 |
0.2 |
GO:1900224 |
positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 |
0.5 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.0 |
1.1 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.0 |
0.2 |
GO:0009052 |
D-ribose metabolic process(GO:0006014) pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 |
0.3 |
GO:0021615 |
glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 |
0.1 |
GO:1990737 |
regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 |
0.5 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 |
0.5 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 |
0.3 |
GO:1902903 |
regulation of fibril organization(GO:1902903) |
| 0.0 |
0.2 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.0 |
0.2 |
GO:0015692 |
lead ion transport(GO:0015692) |
| 0.0 |
0.2 |
GO:0018094 |
protein polyglycylation(GO:0018094) |
| 0.0 |
0.3 |
GO:0032286 |
central nervous system myelin maintenance(GO:0032286) |
| 0.0 |
0.1 |
GO:0042986 |
positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 |
0.3 |
GO:0002759 |
regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 |
0.2 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 |
0.2 |
GO:0050812 |
acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 |
0.3 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.0 |
0.2 |
GO:0016240 |
autophagosome docking(GO:0016240) |
| 0.0 |
0.2 |
GO:2000323 |
negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 |
0.1 |
GO:0043932 |
ossification involved in bone remodeling(GO:0043932) |
| 0.0 |
0.3 |
GO:0051388 |
positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 |
0.5 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.0 |
0.1 |
GO:0007538 |
primary sex determination(GO:0007538) |
| 0.0 |
0.2 |
GO:2000312 |
regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 |
0.1 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 |
0.1 |
GO:0006258 |
UDP-glucose catabolic process(GO:0006258) |
| 0.0 |
0.1 |
GO:1900920 |
regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.0 |
0.1 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.0 |
0.1 |
GO:1900453 |
negative regulation of long term synaptic depression(GO:1900453) |
| 0.0 |
0.1 |
GO:0035359 |
negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 |
0.2 |
GO:0007042 |
lysosomal lumen acidification(GO:0007042) |
| 0.0 |
0.8 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.0 |
0.2 |
GO:0015798 |
myo-inositol transport(GO:0015798) |
| 0.0 |
0.1 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 |
0.1 |
GO:0032240 |
negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 |
0.6 |
GO:0036295 |
cellular response to increased oxygen levels(GO:0036295) |
| 0.0 |
0.2 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.6 |
GO:0072189 |
ureter development(GO:0072189) |
| 0.0 |
0.1 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 |
0.2 |
GO:0032380 |
regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 |
0.8 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.0 |
0.9 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 |
0.3 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.0 |
0.7 |
GO:0072539 |
T-helper 17 cell differentiation(GO:0072539) |
| 0.0 |
0.1 |
GO:0010767 |
regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.0 |
0.0 |
GO:0006288 |
base-excision repair, DNA ligation(GO:0006288) |
| 0.0 |
0.3 |
GO:0045716 |
positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 |
0.1 |
GO:0070427 |
nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 |
0.2 |
GO:0006880 |
intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 |
0.2 |
GO:0051140 |
regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 |
0.1 |
GO:0051029 |
rRNA transport(GO:0051029) |
| 0.0 |
0.1 |
GO:0014050 |
negative regulation of glutamate secretion(GO:0014050) |
| 0.0 |
0.1 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 |
0.1 |
GO:0048069 |
eye pigmentation(GO:0048069) |
| 0.0 |
0.1 |
GO:1901838 |
positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 |
0.2 |
GO:0035751 |
regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 |
0.9 |
GO:0031954 |
positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 |
0.1 |
GO:0000415 |
negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 |
0.1 |
GO:0008063 |
Toll signaling pathway(GO:0008063) |
| 0.0 |
0.1 |
GO:0045065 |
cytotoxic T cell differentiation(GO:0045065) |
| 0.0 |
0.5 |
GO:0000338 |
protein deneddylation(GO:0000338) |
| 0.0 |
0.1 |
GO:0045906 |
negative regulation of vasoconstriction(GO:0045906) |
| 0.0 |
1.7 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.1 |
GO:0071106 |
coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 |
0.9 |
GO:0022400 |
regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 |
1.5 |
GO:0050775 |
positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 |
0.6 |
GO:0021522 |
spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 |
0.4 |
GO:0014029 |
neural crest formation(GO:0014029) |
| 0.0 |
0.2 |
GO:0098712 |
L-glutamate import across plasma membrane(GO:0098712) |
| 0.0 |
0.7 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.0 |
0.3 |
GO:0090199 |
regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 |
0.2 |
GO:1901299 |
negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 |
0.2 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.0 |
0.1 |
GO:0070602 |
regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 |
0.6 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.0 |
2.0 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.0 |
0.1 |
GO:0060754 |
positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 |
1.2 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 |
0.1 |
GO:0002316 |
follicular B cell differentiation(GO:0002316) |
| 0.0 |
0.0 |
GO:0006290 |
pyrimidine dimer repair(GO:0006290) |
| 0.0 |
0.1 |
GO:0001893 |
maternal placenta development(GO:0001893) |
| 0.0 |
0.2 |
GO:0007095 |
mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 |
0.1 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.0 |
0.4 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.4 |
GO:0002726 |
positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 |
0.2 |
GO:0097084 |
vascular smooth muscle cell development(GO:0097084) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 |
0.2 |
GO:0005984 |
disaccharide metabolic process(GO:0005984) |
| 0.0 |
0.1 |
GO:0061052 |
negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 |
0.1 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.0 |
0.4 |
GO:0009125 |
nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 |
0.1 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 |
0.0 |
GO:1904562 |
phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 |
0.2 |
GO:0042908 |
xenobiotic transport(GO:0042908) |
| 0.0 |
0.1 |
GO:2000973 |
regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 |
0.4 |
GO:1902894 |
negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 |
0.1 |
GO:1904617 |
negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 |
0.1 |
GO:0036493 |
positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 |
0.1 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 |
0.4 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.1 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
1.7 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.0 |
0.0 |
GO:2000276 |
negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 |
0.1 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.0 |
0.8 |
GO:0007602 |
phototransduction(GO:0007602) |
| 0.0 |
0.6 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.2 |
GO:0014877 |
response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 |
0.1 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 |
0.1 |
GO:0044387 |
negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 |
0.4 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.0 |
0.3 |
GO:0050872 |
white fat cell differentiation(GO:0050872) |
| 0.0 |
0.4 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
0.1 |
GO:0006424 |
glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 |
0.5 |
GO:1903077 |
negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 |
0.5 |
GO:0070534 |
protein K63-linked ubiquitination(GO:0070534) |
| 0.0 |
0.1 |
GO:0010040 |
response to iron(II) ion(GO:0010040) |
| 0.0 |
0.2 |
GO:0035116 |
embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 |
0.1 |
GO:0045007 |
depurination(GO:0045007) |
| 0.0 |
0.1 |
GO:0046833 |
positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 |
0.8 |
GO:0010862 |
positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 |
0.2 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 |
0.2 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.2 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 |
0.1 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 |
0.0 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 |
0.2 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.0 |
GO:0015917 |
aminophospholipid transport(GO:0015917) |
| 0.0 |
0.1 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 |
0.1 |
GO:0035249 |
synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 |
0.0 |
GO:1903517 |
regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) regulation of single strand break repair(GO:1903516) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 |
0.2 |
GO:0060766 |
negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 |
0.0 |
GO:2000304 |
positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 |
0.2 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.0 |
0.1 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.0 |
0.2 |
GO:0035826 |
rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 |
0.2 |
GO:0016926 |
protein desumoylation(GO:0016926) |
| 0.0 |
0.1 |
GO:0000066 |
mitochondrial ornithine transport(GO:0000066) |
| 0.0 |
0.2 |
GO:0035360 |
positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 |
0.5 |
GO:0006111 |
regulation of gluconeogenesis(GO:0006111) |
| 0.0 |
0.7 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.1 |
GO:0060326 |
cell chemotaxis(GO:0060326) |
| 0.0 |
0.2 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 |
0.1 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 |
0.0 |
GO:0000454 |
snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 |
0.2 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 |
0.3 |
GO:0030828 |
positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 |
0.3 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 |
0.0 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.0 |
0.0 |
GO:0090427 |
monocyte extravasation(GO:0035696) activation of meiosis(GO:0090427) regulation of monocyte extravasation(GO:2000437) |
| 0.0 |
0.1 |
GO:0006552 |
leucine catabolic process(GO:0006552) |
| 0.0 |
0.0 |
GO:0006311 |
meiotic gene conversion(GO:0006311) |
| 0.0 |
0.1 |
GO:0055129 |
L-proline biosynthetic process(GO:0055129) |
| 0.0 |
0.0 |
GO:0060510 |
Type II pneumocyte differentiation(GO:0060510) |
| 0.0 |
0.0 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 |
0.1 |
GO:0051342 |
regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 |
0.1 |
GO:0061469 |
regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 |
0.2 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.0 |
GO:0032223 |
negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 |
0.0 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 |
0.1 |
GO:0016574 |
histone ubiquitination(GO:0016574) |
| 0.0 |
0.1 |
GO:1903206 |
negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.0 |
0.0 |
GO:0002143 |
tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 |
0.1 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.0 |
0.1 |
GO:0035019 |
somatic stem cell population maintenance(GO:0035019) |
| 0.0 |
0.0 |
GO:0042350 |
GDP-L-fucose biosynthetic process(GO:0042350) |
| 0.0 |
0.3 |
GO:0007130 |
synaptonemal complex assembly(GO:0007130) |
| 0.0 |
0.3 |
GO:0035455 |
response to interferon-alpha(GO:0035455) |
| 0.0 |
0.1 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.1 |
GO:1904714 |
regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 |
0.1 |
GO:0015781 |
pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 |
0.1 |
GO:0032287 |
peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 |
0.0 |
GO:0060127 |
subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung development(GO:0060459) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) endodermal digestive tract morphogenesis(GO:0061031) |
| 0.0 |
0.3 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 |
0.1 |
GO:0080182 |
histone H3-K4 trimethylation(GO:0080182) |
| 0.0 |
0.3 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.1 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.0 |
0.1 |
GO:0051934 |
dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 |
0.1 |
GO:0034244 |
positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.2 |
GO:0042737 |
drug catabolic process(GO:0042737) |
| 0.0 |
0.1 |
GO:0033005 |
positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 |
0.2 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.1 |
GO:0045070 |
positive regulation of viral genome replication(GO:0045070) |
| 0.0 |
0.1 |
GO:1902570 |
protein localization to nucleolus(GO:1902570) |
| 0.0 |
0.2 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 |
0.4 |
GO:0009267 |
cellular response to starvation(GO:0009267) |
| 0.0 |
1.1 |
GO:0006521 |
regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 |
0.1 |
GO:0097091 |
synaptic vesicle clustering(GO:0097091) |
| 0.0 |
0.1 |
GO:2000042 |
negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 |
0.9 |
GO:0097031 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.2 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.0 |
0.1 |
GO:0031087 |
deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 |
0.1 |
GO:0070911 |
global genome nucleotide-excision repair(GO:0070911) |
| 0.0 |
0.9 |
GO:0030433 |
ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 |
0.2 |
GO:0060088 |
auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 |
0.1 |
GO:0016554 |
cytidine to uridine editing(GO:0016554) |