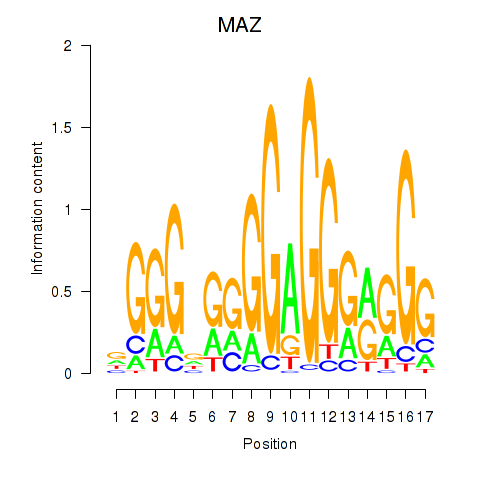
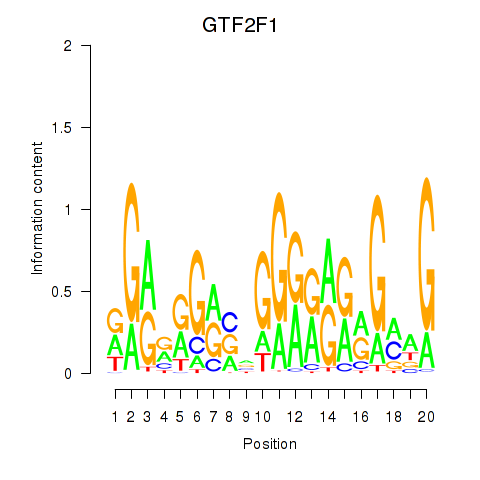
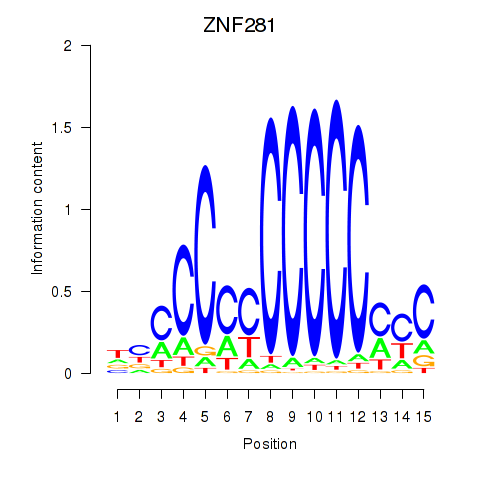
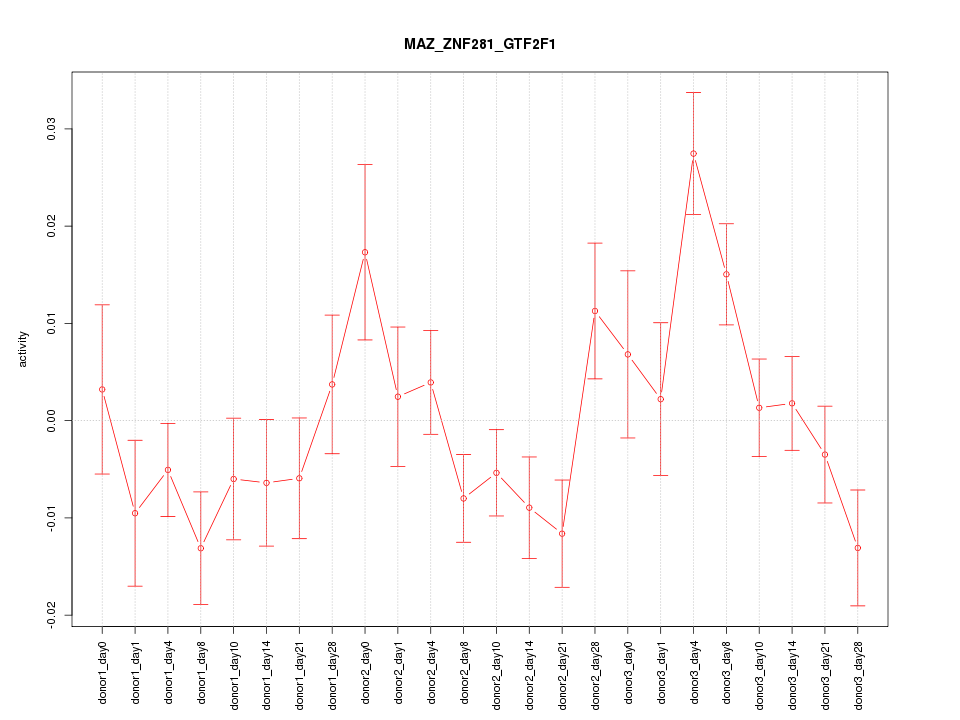
| 2.0 |
12.1 |
GO:0002803 |
positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 2.0 |
6.0 |
GO:0046586 |
regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 1.1 |
3.2 |
GO:0060516 |
primary prostatic bud elongation(GO:0060516) |
| 1.0 |
3.0 |
GO:2001037 |
tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 1.0 |
2.9 |
GO:0031630 |
regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.8 |
2.5 |
GO:0071109 |
superior temporal gyrus development(GO:0071109) |
| 0.8 |
2.5 |
GO:0052047 |
interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.8 |
2.4 |
GO:0009946 |
proximal/distal axis specification(GO:0009946) |
| 0.8 |
4.0 |
GO:0021773 |
striatal medium spiny neuron differentiation(GO:0021773) |
| 0.7 |
6.0 |
GO:0031444 |
slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.6 |
0.6 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.6 |
3.2 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.6 |
0.6 |
GO:0050965 |
detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.6 |
1.7 |
GO:1904298 |
positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 0.5 |
1.6 |
GO:0006535 |
cysteine biosynthetic process from serine(GO:0006535) |
| 0.5 |
2.2 |
GO:0070384 |
Harderian gland development(GO:0070384) |
| 0.5 |
1.6 |
GO:0050894 |
determination of affect(GO:0050894) |
| 0.5 |
1.6 |
GO:1901899 |
positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.5 |
2.1 |
GO:0033123 |
positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.5 |
1.0 |
GO:0031133 |
regulation of axon diameter(GO:0031133) |
| 0.5 |
11.2 |
GO:0016540 |
protein autoprocessing(GO:0016540) |
| 0.5 |
4.5 |
GO:0070236 |
negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.5 |
2.5 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 0.5 |
1.5 |
GO:0021938 |
ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.5 |
1.4 |
GO:0033242 |
regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.5 |
2.9 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.5 |
3.8 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.5 |
2.3 |
GO:0010609 |
mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.5 |
0.5 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.5 |
1.4 |
GO:1903519 |
apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.5 |
2.3 |
GO:0048749 |
compound eye development(GO:0048749) |
| 0.4 |
1.8 |
GO:0003404 |
optic vesicle morphogenesis(GO:0003404) optic cup structural organization(GO:0003409) |
| 0.4 |
1.3 |
GO:0015920 |
lipopolysaccharide transport(GO:0015920) |
| 0.4 |
1.3 |
GO:0061386 |
closure of optic fissure(GO:0061386) |
| 0.4 |
1.7 |
GO:0010752 |
regulation of cGMP-mediated signaling(GO:0010752) |
| 0.4 |
0.4 |
GO:0001952 |
regulation of cell-matrix adhesion(GO:0001952) |
| 0.4 |
1.2 |
GO:0003050 |
regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.4 |
2.0 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.4 |
1.6 |
GO:1905167 |
positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.4 |
1.9 |
GO:1902378 |
vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.4 |
3.5 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.4 |
1.2 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.4 |
1.1 |
GO:0090271 |
positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.4 |
1.5 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.4 |
1.1 |
GO:0001300 |
chronological cell aging(GO:0001300) |
| 0.4 |
0.4 |
GO:1904468 |
negative regulation of tumor necrosis factor secretion(GO:1904468) |
| 0.4 |
0.4 |
GO:1904862 |
inhibitory synapse assembly(GO:1904862) |
| 0.4 |
1.4 |
GO:0021650 |
vestibulocochlear nerve formation(GO:0021650) |
| 0.4 |
1.4 |
GO:0018352 |
protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.3 |
1.0 |
GO:0021798 |
forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.3 |
1.7 |
GO:0061143 |
alveolar primary septum development(GO:0061143) |
| 0.3 |
2.4 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.3 |
0.7 |
GO:2000646 |
positive regulation of receptor catabolic process(GO:2000646) |
| 0.3 |
1.0 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 0.3 |
1.7 |
GO:0071802 |
negative regulation of podosome assembly(GO:0071802) |
| 0.3 |
4.2 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.3 |
1.0 |
GO:0017186 |
peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.3 |
3.6 |
GO:0034465 |
response to carbon monoxide(GO:0034465) |
| 0.3 |
0.6 |
GO:0060557 |
positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.3 |
1.3 |
GO:1902723 |
negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.3 |
1.3 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.3 |
1.3 |
GO:0032912 |
negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.3 |
0.9 |
GO:0043449 |
cellular alkene metabolic process(GO:0043449) |
| 0.3 |
1.5 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.3 |
3.7 |
GO:0035986 |
senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.3 |
0.6 |
GO:1903422 |
negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.3 |
0.9 |
GO:0030264 |
nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.3 |
2.7 |
GO:0097116 |
postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.3 |
0.9 |
GO:0021526 |
medial motor column neuron differentiation(GO:0021526) |
| 0.3 |
1.5 |
GO:1904674 |
positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.3 |
0.9 |
GO:0097254 |
renal tubular secretion(GO:0097254) |
| 0.3 |
1.5 |
GO:0032455 |
nerve growth factor processing(GO:0032455) |
| 0.3 |
2.1 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 |
0.3 |
GO:2000724 |
positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.3 |
0.3 |
GO:0090170 |
regulation of Golgi inheritance(GO:0090170) |
| 0.3 |
0.9 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.3 |
1.5 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.3 |
2.9 |
GO:0060020 |
Bergmann glial cell differentiation(GO:0060020) |
| 0.3 |
0.9 |
GO:0032499 |
detection of peptidoglycan(GO:0032499) |
| 0.3 |
0.9 |
GO:0000412 |
histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.3 |
1.1 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.3 |
3.7 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.3 |
7.4 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.3 |
0.9 |
GO:0018057 |
peptidyl-lysine oxidation(GO:0018057) |
| 0.3 |
0.9 |
GO:0090187 |
positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.3 |
2.5 |
GO:1902746 |
regulation of lens fiber cell differentiation(GO:1902746) |
| 0.3 |
0.8 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 |
0.3 |
GO:0014835 |
myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of anagen(GO:0051885) |
| 0.3 |
0.3 |
GO:0048262 |
determination of dorsal/ventral asymmetry(GO:0048262) determination of dorsal identity(GO:0048263) |
| 0.3 |
1.9 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.3 |
0.8 |
GO:0003169 |
coronary vein morphogenesis(GO:0003169) regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.3 |
3.0 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 |
0.8 |
GO:0001172 |
transcription, RNA-templated(GO:0001172) |
| 0.3 |
1.1 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 |
0.8 |
GO:0051780 |
mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.3 |
0.3 |
GO:0016199 |
axon midline choice point recognition(GO:0016199) |
| 0.3 |
1.1 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 |
1.0 |
GO:0021603 |
cranial nerve formation(GO:0021603) |
| 0.3 |
0.8 |
GO:0021965 |
spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.3 |
0.8 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.3 |
1.3 |
GO:0003322 |
pancreatic A cell development(GO:0003322) |
| 0.3 |
1.0 |
GO:1903691 |
positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.2 |
1.0 |
GO:0071477 |
hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.2 |
0.5 |
GO:2000309 |
activation of MAPK activity involved in innate immune response(GO:0035419) positive regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000309) |
| 0.2 |
1.2 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 |
1.5 |
GO:0009082 |
branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 |
1.5 |
GO:0051012 |
microtubule sliding(GO:0051012) |
| 0.2 |
0.7 |
GO:0048627 |
myoblast development(GO:0048627) |
| 0.2 |
2.4 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.2 |
2.9 |
GO:0002741 |
positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.2 |
1.4 |
GO:0070141 |
response to UV-A(GO:0070141) |
| 0.2 |
0.2 |
GO:0060157 |
urinary bladder development(GO:0060157) |
| 0.2 |
1.4 |
GO:0086098 |
angiotensin-activated signaling pathway involved in heart process(GO:0086098) |
| 0.2 |
0.9 |
GO:0010900 |
regulation of phosphatidylcholine catabolic process(GO:0010899) negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.2 |
1.9 |
GO:0010767 |
regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.2 |
1.8 |
GO:0070120 |
ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.2 |
1.3 |
GO:0030421 |
defecation(GO:0030421) |
| 0.2 |
0.7 |
GO:0019072 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 |
1.5 |
GO:0021957 |
corticospinal tract morphogenesis(GO:0021957) |
| 0.2 |
0.7 |
GO:0045226 |
extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 |
0.7 |
GO:0014707 |
branchiomeric skeletal muscle development(GO:0014707) |
| 0.2 |
0.4 |
GO:0021859 |
pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.2 |
0.9 |
GO:0033159 |
negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.2 |
0.6 |
GO:0045210 |
FasL biosynthetic process(GO:0045210) |
| 0.2 |
0.6 |
GO:1990869 |
response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.2 |
0.9 |
GO:1901490 |
regulation of lymphangiogenesis(GO:1901490) |
| 0.2 |
0.2 |
GO:0016572 |
histone phosphorylation(GO:0016572) |
| 0.2 |
0.4 |
GO:0003166 |
bundle of His development(GO:0003166) |
| 0.2 |
0.6 |
GO:0048250 |
mitochondrial iron ion transport(GO:0048250) |
| 0.2 |
1.9 |
GO:1902866 |
regulation of retina development in camera-type eye(GO:1902866) |
| 0.2 |
1.2 |
GO:0000432 |
regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.2 |
0.8 |
GO:0042494 |
detection of bacterial lipoprotein(GO:0042494) |
| 0.2 |
0.4 |
GO:0007161 |
calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.2 |
0.4 |
GO:0086048 |
membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.2 |
1.0 |
GO:1903377 |
negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.2 |
3.1 |
GO:0010867 |
positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.2 |
0.8 |
GO:0035441 |
cell migration involved in vasculogenesis(GO:0035441) |
| 0.2 |
0.6 |
GO:2000910 |
negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.2 |
0.4 |
GO:1903862 |
positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.2 |
1.4 |
GO:0021842 |
directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.2 |
0.2 |
GO:0035793 |
negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.2 |
0.2 |
GO:1902938 |
regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.2 |
1.6 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.2 |
3.2 |
GO:0030949 |
positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.2 |
7.8 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.2 |
0.8 |
GO:2000686 |
regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.2 |
3.6 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.2 |
0.2 |
GO:0097084 |
vascular smooth muscle cell development(GO:0097084) |
| 0.2 |
0.8 |
GO:2000286 |
receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.2 |
1.0 |
GO:0051122 |
hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 |
4.1 |
GO:0001502 |
cartilage condensation(GO:0001502) |
| 0.2 |
1.5 |
GO:0007506 |
gonadal mesoderm development(GO:0007506) |
| 0.2 |
0.6 |
GO:1900042 |
positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 |
0.8 |
GO:0060426 |
lung vasculature development(GO:0060426) |
| 0.2 |
1.1 |
GO:1902714 |
negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.2 |
1.5 |
GO:0003065 |
positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.2 |
1.0 |
GO:0007070 |
negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.2 |
1.5 |
GO:0048050 |
post-embryonic eye morphogenesis(GO:0048050) |
| 0.2 |
0.6 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.2 |
0.9 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.2 |
0.2 |
GO:0060585 |
regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.2 |
0.6 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 |
0.4 |
GO:0003192 |
mitral valve formation(GO:0003192) |
| 0.2 |
0.7 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.2 |
0.4 |
GO:0090107 |
regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.2 |
0.2 |
GO:1903895 |
negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.2 |
0.7 |
GO:0032487 |
regulation of Rap protein signal transduction(GO:0032487) |
| 0.2 |
0.5 |
GO:1904328 |
regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.2 |
0.7 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.2 |
1.6 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.2 |
1.8 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 |
0.7 |
GO:0097010 |
eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.2 |
0.2 |
GO:1903588 |
negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.2 |
0.7 |
GO:0097498 |
endothelial tube lumen extension(GO:0097498) |
| 0.2 |
1.2 |
GO:2000587 |
negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.2 |
2.4 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 |
0.5 |
GO:0045658 |
regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.2 |
0.3 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.2 |
0.7 |
GO:0036269 |
swimming behavior(GO:0036269) |
| 0.2 |
1.0 |
GO:0071895 |
odontoblast differentiation(GO:0071895) |
| 0.2 |
0.5 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.2 |
0.2 |
GO:2000611 |
positive regulation of thyroid hormone generation(GO:2000611) |
| 0.2 |
1.0 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.2 |
0.2 |
GO:0045939 |
negative regulation of steroid biosynthetic process(GO:0010894) negative regulation of steroid metabolic process(GO:0045939) |
| 0.2 |
0.2 |
GO:0035413 |
positive regulation of catenin import into nucleus(GO:0035413) |
| 0.2 |
0.5 |
GO:0021503 |
neural fold bending(GO:0021503) |
| 0.2 |
1.7 |
GO:0048711 |
positive regulation of astrocyte differentiation(GO:0048711) |
| 0.2 |
0.3 |
GO:0042635 |
positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.2 |
1.7 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.2 |
2.5 |
GO:1904778 |
regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 |
0.7 |
GO:2000342 |
negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.2 |
0.2 |
GO:0021626 |
hindbrain maturation(GO:0021578) central nervous system maturation(GO:0021626) superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.2 |
0.2 |
GO:2000196 |
positive regulation of female gonad development(GO:2000196) |
| 0.2 |
0.6 |
GO:0010760 |
negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.2 |
0.5 |
GO:0098937 |
dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.2 |
0.6 |
GO:0014054 |
positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.2 |
1.0 |
GO:0010868 |
negative regulation of triglyceride biosynthetic process(GO:0010868) |
| 0.2 |
1.4 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 |
0.3 |
GO:0061073 |
ciliary body morphogenesis(GO:0061073) |
| 0.2 |
0.6 |
GO:0010579 |
regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.2 |
0.6 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.2 |
0.6 |
GO:1900168 |
glial cell-derived neurotrophic factor secretion(GO:0044467) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.2 |
0.5 |
GO:0050919 |
negative chemotaxis(GO:0050919) |
| 0.2 |
1.4 |
GO:0046958 |
nonassociative learning(GO:0046958) |
| 0.2 |
0.5 |
GO:0042361 |
menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.2 |
0.3 |
GO:0060290 |
transdifferentiation(GO:0060290) |
| 0.2 |
0.2 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 |
0.8 |
GO:1900425 |
negative regulation of defense response to bacterium(GO:1900425) |
| 0.2 |
0.6 |
GO:0014028 |
notochord formation(GO:0014028) |
| 0.2 |
1.8 |
GO:0060068 |
vagina development(GO:0060068) |
| 0.1 |
4.0 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 |
0.3 |
GO:0060019 |
radial glial cell differentiation(GO:0060019) |
| 0.1 |
0.4 |
GO:0035544 |
negative regulation of SNARE complex assembly(GO:0035544) |
| 0.1 |
0.7 |
GO:0003104 |
positive regulation of glomerular filtration(GO:0003104) |
| 0.1 |
0.4 |
GO:0030186 |
melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 |
0.1 |
GO:0019233 |
sensory perception of pain(GO:0019233) |
| 0.1 |
0.6 |
GO:0072069 |
DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) |
| 0.1 |
0.6 |
GO:0046086 |
adenosine biosynthetic process(GO:0046086) |
| 0.1 |
1.6 |
GO:0090009 |
primitive streak formation(GO:0090009) |
| 0.1 |
0.4 |
GO:0070563 |
negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 |
0.6 |
GO:0030886 |
negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.1 |
0.4 |
GO:1904604 |
regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904596) negative regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904597) regulation of advanced glycation end-product receptor activity(GO:1904603) negative regulation of advanced glycation end-product receptor activity(GO:1904604) negative regulation of connective tissue replacement(GO:1905204) |
| 0.1 |
0.3 |
GO:0097187 |
dentinogenesis(GO:0097187) |
| 0.1 |
0.7 |
GO:1904640 |
response to methionine(GO:1904640) |
| 0.1 |
0.4 |
GO:0060148 |
positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 |
0.3 |
GO:0031247 |
actin rod assembly(GO:0031247) |
| 0.1 |
0.8 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
1.1 |
GO:0051126 |
negative regulation of actin nucleation(GO:0051126) |
| 0.1 |
0.3 |
GO:0021812 |
neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 |
1.2 |
GO:0035860 |
glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 |
0.3 |
GO:0021590 |
cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.1 |
0.5 |
GO:2001013 |
epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 |
1.4 |
GO:0030903 |
notochord development(GO:0030903) |
| 0.1 |
0.4 |
GO:0099525 |
presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 |
1.6 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 |
0.1 |
GO:0097421 |
liver regeneration(GO:0097421) |
| 0.1 |
1.5 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 |
0.7 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 |
0.1 |
GO:2000777 |
positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 |
0.1 |
GO:1901860 |
positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.1 |
1.3 |
GO:1904417 |
regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.1 |
1.2 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
1.2 |
GO:0045636 |
positive regulation of melanocyte differentiation(GO:0045636) |
| 0.1 |
0.7 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.1 |
0.3 |
GO:0046532 |
regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 |
0.4 |
GO:0001207 |
histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 |
0.4 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.1 |
0.4 |
GO:2000824 |
negative regulation of androgen receptor activity(GO:2000824) |
| 0.1 |
0.3 |
GO:0071484 |
cellular response to light intensity(GO:0071484) |
| 0.1 |
0.1 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 |
0.3 |
GO:0003308 |
negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.1 |
2.5 |
GO:0030575 |
nuclear body organization(GO:0030575) |
| 0.1 |
0.5 |
GO:0043376 |
regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 |
0.5 |
GO:2000852 |
corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.1 |
1.0 |
GO:1903764 |
regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 |
0.9 |
GO:0051461 |
positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 |
1.7 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.1 |
0.1 |
GO:0048733 |
sebaceous gland development(GO:0048733) |
| 0.1 |
0.6 |
GO:1990834 |
response to odorant(GO:1990834) |
| 0.1 |
0.5 |
GO:0006565 |
L-serine catabolic process(GO:0006565) |
| 0.1 |
0.9 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.1 |
0.4 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 |
0.1 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) |
| 0.1 |
0.4 |
GO:0038165 |
oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 |
1.2 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 |
1.9 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.1 |
0.9 |
GO:0060753 |
regulation of mast cell chemotaxis(GO:0060753) |
| 0.1 |
0.4 |
GO:1990258 |
box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.1 |
0.2 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 |
0.5 |
GO:0018242 |
protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 |
1.0 |
GO:1904627 |
response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 |
0.2 |
GO:0020027 |
hemoglobin metabolic process(GO:0020027) |
| 0.1 |
0.1 |
GO:1903923 |
protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 |
0.8 |
GO:1904179 |
positive regulation of adipose tissue development(GO:1904179) |
| 0.1 |
1.4 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
0.4 |
GO:0046947 |
hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 |
1.2 |
GO:0098734 |
macromolecule depalmitoylation(GO:0098734) |
| 0.1 |
0.1 |
GO:2000765 |
regulation of cytoplasmic translation(GO:2000765) |
| 0.1 |
0.4 |
GO:0061534 |
gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 |
0.6 |
GO:0090324 |
negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.1 |
6.1 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 |
0.5 |
GO:0032185 |
septin cytoskeleton organization(GO:0032185) |
| 0.1 |
0.1 |
GO:1903181 |
regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.1 |
2.8 |
GO:0097062 |
dendritic spine maintenance(GO:0097062) |
| 0.1 |
0.5 |
GO:0038026 |
reelin-mediated signaling pathway(GO:0038026) |
| 0.1 |
2.1 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 |
0.2 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 |
1.2 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 |
2.0 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.1 |
8.7 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.1 |
0.3 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.1 |
0.5 |
GO:0033686 |
positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.1 |
0.1 |
GO:0036480 |
neuron intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0036480) regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903376) |
| 0.1 |
2.4 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.1 |
0.1 |
GO:0010982 |
regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 |
0.1 |
GO:0014048 |
regulation of glutamate secretion(GO:0014048) |
| 0.1 |
0.3 |
GO:2000969 |
positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 |
0.1 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 |
0.6 |
GO:0072679 |
thymocyte migration(GO:0072679) |
| 0.1 |
0.2 |
GO:1902713 |
regulation of interferon-gamma secretion(GO:1902713) |
| 0.1 |
0.7 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 |
0.3 |
GO:0097069 |
cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.1 |
0.6 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.1 |
1.3 |
GO:0001542 |
ovulation from ovarian follicle(GO:0001542) |
| 0.1 |
0.1 |
GO:0097051 |
establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 |
1.2 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.1 |
0.7 |
GO:0010642 |
negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.1 |
0.3 |
GO:0007538 |
primary sex determination(GO:0007538) |
| 0.1 |
0.5 |
GO:0014050 |
negative regulation of glutamate secretion(GO:0014050) |
| 0.1 |
0.2 |
GO:0086092 |
regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.1 |
0.1 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.1 |
1.0 |
GO:0032776 |
DNA methylation on cytosine(GO:0032776) |
| 0.1 |
0.4 |
GO:2000418 |
positive regulation of eosinophil migration(GO:2000418) |
| 0.1 |
0.9 |
GO:0035524 |
proline transmembrane transport(GO:0035524) |
| 0.1 |
0.4 |
GO:0045602 |
negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 |
1.8 |
GO:0001845 |
phagolysosome assembly(GO:0001845) |
| 0.1 |
0.1 |
GO:0010891 |
negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 |
1.0 |
GO:0098712 |
L-glutamate import across plasma membrane(GO:0098712) |
| 0.1 |
1.3 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 |
0.5 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 |
0.1 |
GO:0021937 |
cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 |
0.6 |
GO:0098707 |
ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 |
1.3 |
GO:0015816 |
glycine transport(GO:0015816) |
| 0.1 |
0.1 |
GO:0098828 |
positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 |
0.8 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.5 |
GO:0042421 |
norepinephrine biosynthetic process(GO:0042421) |
| 0.1 |
0.3 |
GO:0038003 |
opioid receptor signaling pathway(GO:0038003) |
| 0.1 |
0.2 |
GO:0071105 |
response to interleukin-11(GO:0071105) |
| 0.1 |
0.6 |
GO:0070458 |
cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 |
0.3 |
GO:1900224 |
positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 |
0.8 |
GO:0035583 |
sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 |
0.1 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 |
1.2 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 |
0.3 |
GO:0031291 |
Ran protein signal transduction(GO:0031291) |
| 0.1 |
0.2 |
GO:0014878 |
response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 |
0.1 |
GO:0002522 |
leukocyte migration involved in immune response(GO:0002522) |
| 0.1 |
0.3 |
GO:0002072 |
optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.1 |
0.8 |
GO:0033210 |
leptin-mediated signaling pathway(GO:0033210) |
| 0.1 |
0.3 |
GO:0002644 |
negative regulation of tolerance induction(GO:0002644) |
| 0.1 |
1.1 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.1 |
0.5 |
GO:0030241 |
skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 |
0.3 |
GO:0042700 |
luteinizing hormone signaling pathway(GO:0042700) |
| 0.1 |
0.1 |
GO:0032707 |
negative regulation of interleukin-23 production(GO:0032707) |
| 0.1 |
0.3 |
GO:0035623 |
regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.1 |
1.9 |
GO:1901897 |
regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 |
0.6 |
GO:1903799 |
negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.1 |
0.5 |
GO:0032525 |
somite rostral/caudal axis specification(GO:0032525) |
| 0.1 |
1.3 |
GO:0036066 |
protein O-linked fucosylation(GO:0036066) |
| 0.1 |
0.8 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 |
0.1 |
GO:2001184 |
positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 |
0.7 |
GO:0055096 |
lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 |
0.2 |
GO:0033563 |
dorsal/ventral axon guidance(GO:0033563) |
| 0.1 |
0.3 |
GO:1904058 |
positive regulation of sensory perception of pain(GO:1904058) |
| 0.1 |
0.2 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.1 |
0.2 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 |
0.7 |
GO:0086045 |
membrane depolarization during AV node cell action potential(GO:0086045) membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.1 |
0.7 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.1 |
0.8 |
GO:2000553 |
positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.1 |
0.5 |
GO:0060356 |
leucine transport(GO:0015820) leucine import(GO:0060356) |
| 0.1 |
2.9 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.1 |
0.3 |
GO:0051901 |
positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.1 |
1.0 |
GO:0010917 |
negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.1 |
0.7 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.1 |
0.8 |
GO:0045741 |
positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 |
1.4 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 |
0.4 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.1 |
0.1 |
GO:0010482 |
epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 |
0.3 |
GO:0034445 |
regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.1 |
3.6 |
GO:1903861 |
positive regulation of dendrite extension(GO:1903861) |
| 0.1 |
0.2 |
GO:1902474 |
positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 |
0.5 |
GO:0035610 |
protein side chain deglutamylation(GO:0035610) |
| 0.1 |
0.1 |
GO:0042398 |
cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.1 |
0.3 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
0.4 |
GO:0090649 |
response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 |
0.3 |
GO:0019285 |
glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 |
2.9 |
GO:0014046 |
dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 |
0.2 |
GO:0021571 |
rhombomere 5 development(GO:0021571) |
| 0.1 |
0.6 |
GO:2000504 |
positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 |
3.0 |
GO:0030207 |
chondroitin sulfate catabolic process(GO:0030207) |
| 0.1 |
0.3 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 |
0.3 |
GO:0090259 |
regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.1 |
0.3 |
GO:0098838 |
reduced folate transmembrane transport(GO:0098838) |
| 0.1 |
0.2 |
GO:0035054 |
embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.1 |
0.8 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.1 |
0.4 |
GO:0017198 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 |
0.6 |
GO:0006021 |
inositol biosynthetic process(GO:0006021) |
| 0.1 |
0.5 |
GO:0044208 |
'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 |
0.5 |
GO:0036015 |
response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.1 |
0.1 |
GO:1904783 |
positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.1 |
1.1 |
GO:0099638 |
endosome to plasma membrane protein transport(GO:0099638) |
| 0.1 |
0.5 |
GO:0038031 |
non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 |
0.5 |
GO:0060613 |
fat pad development(GO:0060613) |
| 0.1 |
0.5 |
GO:0050917 |
sensory perception of umami taste(GO:0050917) |
| 0.1 |
0.5 |
GO:0019346 |
homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.1 |
0.5 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 |
0.4 |
GO:0030261 |
chromosome condensation(GO:0030261) |
| 0.1 |
0.5 |
GO:0052551 |
response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.1 |
0.3 |
GO:0042369 |
vitamin D catabolic process(GO:0042369) |
| 0.1 |
0.1 |
GO:1900452 |
regulation of long term synaptic depression(GO:1900452) |
| 0.1 |
3.1 |
GO:0097503 |
sialylation(GO:0097503) |
| 0.1 |
0.4 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 |
0.4 |
GO:0038195 |
urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 |
2.1 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.1 |
0.3 |
GO:0033693 |
neurofilament bundle assembly(GO:0033693) |
| 0.1 |
0.4 |
GO:0050968 |
detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 |
0.4 |
GO:0071474 |
cellular hyperosmotic response(GO:0071474) |
| 0.1 |
1.2 |
GO:0031580 |
membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 |
0.3 |
GO:0060084 |
synaptic transmission involved in micturition(GO:0060084) |
| 0.1 |
0.3 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 |
0.4 |
GO:1903969 |
mammary gland fat development(GO:0060611) regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) regulation of response to macrophage colony-stimulating factor(GO:1903969) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.1 |
0.3 |
GO:0003095 |
pressure natriuresis(GO:0003095) |
| 0.1 |
0.4 |
GO:1902499 |
positive regulation of protein autoubiquitination(GO:1902499) |
| 0.1 |
0.4 |
GO:0072526 |
pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 |
0.1 |
GO:0010986 |
positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 |
0.2 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.1 |
0.6 |
GO:1904885 |
beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 |
0.2 |
GO:0021562 |
vestibulocochlear nerve development(GO:0021562) |
| 0.1 |
0.3 |
GO:0071205 |
protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 |
0.1 |
GO:0043006 |
activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 |
0.2 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 |
3.2 |
GO:0046627 |
negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.1 |
0.3 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 |
1.0 |
GO:0050884 |
neuromuscular process controlling posture(GO:0050884) |
| 0.1 |
0.2 |
GO:0043397 |
corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.1 |
0.1 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.1 |
0.5 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 |
0.3 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.1 |
0.8 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 |
0.3 |
GO:0043653 |
mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 |
0.3 |
GO:0044828 |
negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 |
0.2 |
GO:0014826 |
vein smooth muscle contraction(GO:0014826) |
| 0.1 |
0.2 |
GO:0003415 |
chondrocyte hypertrophy(GO:0003415) |
| 0.1 |
0.2 |
GO:0010897 |
negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.1 |
0.3 |
GO:0070561 |
vitamin D receptor signaling pathway(GO:0070561) |
| 0.1 |
0.2 |
GO:0070167 |
regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 |
0.4 |
GO:1904995 |
negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.1 |
0.3 |
GO:0032484 |
Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 |
0.9 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.1 |
0.4 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 |
0.1 |
GO:0061373 |
mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) |
| 0.1 |
0.2 |
GO:1902731 |
negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 |
0.1 |
GO:0036065 |
fucosylation(GO:0036065) |
| 0.1 |
0.5 |
GO:0050910 |
detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 |
0.2 |
GO:0002042 |
cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.1 |
0.2 |
GO:0060484 |
lung-associated mesenchyme development(GO:0060484) |
| 0.1 |
0.9 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 |
0.3 |
GO:0034224 |
cellular response to zinc ion starvation(GO:0034224) |
| 0.1 |
0.2 |
GO:0002881 |
negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.1 |
0.6 |
GO:0034315 |
regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 |
0.4 |
GO:0021546 |
rhombomere development(GO:0021546) |
| 0.1 |
0.9 |
GO:0021891 |
olfactory bulb interneuron development(GO:0021891) |
| 0.1 |
0.5 |
GO:0070970 |
interleukin-2 secretion(GO:0070970) |
| 0.1 |
0.2 |
GO:0007185 |
transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 |
0.2 |
GO:0035624 |
receptor transactivation(GO:0035624) |
| 0.1 |
0.2 |
GO:0010841 |
positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.1 |
0.4 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 |
0.8 |
GO:0090129 |
positive regulation of synapse maturation(GO:0090129) |
| 0.1 |
0.1 |
GO:0072257 |
metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 |
0.1 |
GO:0060935 |
cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) |
| 0.1 |
0.2 |
GO:0019417 |
sulfur oxidation(GO:0019417) |
| 0.1 |
0.7 |
GO:0086073 |
bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 |
0.4 |
GO:0045019 |
negative regulation of nitric oxide biosynthetic process(GO:0045019) negative regulation of nitric oxide metabolic process(GO:1904406) |
| 0.1 |
0.2 |
GO:0018008 |
N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.1 |
0.4 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 |
0.4 |
GO:0019344 |
cysteine biosynthetic process(GO:0019344) |
| 0.1 |
0.5 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 |
0.8 |
GO:0009629 |
response to gravity(GO:0009629) |
| 0.1 |
0.9 |
GO:0010839 |
negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 |
1.4 |
GO:0032196 |
transposition(GO:0032196) |
| 0.1 |
0.2 |
GO:0002276 |
basophil activation involved in immune response(GO:0002276) |
| 0.1 |
0.3 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.1 |
0.1 |
GO:0034163 |
regulation of toll-like receptor 9 signaling pathway(GO:0034163) |
| 0.1 |
0.3 |
GO:0030240 |
skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 |
0.1 |
GO:0010716 |
negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.1 |
0.2 |
GO:0002636 |
positive regulation of germinal center formation(GO:0002636) |
| 0.1 |
0.4 |
GO:2000312 |
regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 |
0.7 |
GO:0036155 |
acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.1 |
0.5 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 |
0.5 |
GO:0097296 |
activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 |
0.4 |
GO:0060215 |
primitive hemopoiesis(GO:0060215) |
| 0.1 |
0.9 |
GO:0030277 |
maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 |
1.9 |
GO:0006509 |
membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 |
0.1 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 |
1.1 |
GO:0048265 |
response to pain(GO:0048265) |
| 0.1 |
0.1 |
GO:0032661 |
regulation of interleukin-18 production(GO:0032661) |
| 0.1 |
0.3 |
GO:2001238 |
positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 |
0.1 |
GO:0048861 |
leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 |
0.1 |
GO:0090118 |
receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.1 |
0.4 |
GO:2000546 |
positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 |
1.4 |
GO:2000811 |
negative regulation of anoikis(GO:2000811) |
| 0.1 |
0.4 |
GO:0044878 |
mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 |
0.2 |
GO:0060178 |
regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.1 |
0.2 |
GO:0050923 |
regulation of negative chemotaxis(GO:0050923) |
| 0.1 |
0.4 |
GO:0039663 |
fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 |
0.1 |
GO:1990637 |
response to prolactin(GO:1990637) |
| 0.1 |
0.3 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 |
0.1 |
GO:0086091 |
regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.1 |
1.0 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.1 |
0.1 |
GO:0050902 |
leukocyte adhesive activation(GO:0050902) |
| 0.1 |
0.2 |
GO:0046166 |
glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 |
0.6 |
GO:1900029 |
positive regulation of ruffle assembly(GO:1900029) |
| 0.1 |
0.1 |
GO:1990641 |
response to iron ion starvation(GO:1990641) |
| 0.1 |
0.6 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 |
0.1 |
GO:0060699 |
regulation of endoribonuclease activity(GO:0060699) |
| 0.1 |
0.2 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 |
1.0 |
GO:0060394 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 |
0.2 |
GO:1904717 |
excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 |
0.2 |
GO:0061107 |
seminal vesicle development(GO:0061107) |
| 0.1 |
0.7 |
GO:0051024 |
positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.1 |
0.5 |
GO:0014877 |
response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 |
0.1 |
GO:0014858 |
positive regulation of skeletal muscle cell proliferation(GO:0014858) |
| 0.1 |
0.1 |
GO:0042159 |
lipoprotein catabolic process(GO:0042159) |
| 0.1 |
0.1 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.1 |
0.3 |
GO:0045663 |
positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 |
0.1 |
GO:0061042 |
vascular wound healing(GO:0061042) |
| 0.1 |
0.2 |
GO:0016095 |
polyprenol catabolic process(GO:0016095) |
| 0.1 |
0.3 |
GO:0040033 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 |
0.7 |
GO:0042178 |
xenobiotic catabolic process(GO:0042178) |
| 0.1 |
0.4 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 |
1.4 |
GO:1901642 |
nucleoside transmembrane transport(GO:1901642) |
| 0.1 |
0.1 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 |
0.1 |
GO:0001560 |
regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 |
0.4 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 |
0.3 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 |
0.4 |
GO:0051026 |
chiasma assembly(GO:0051026) |
| 0.1 |
1.5 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 |
0.3 |
GO:0048318 |
axial mesoderm development(GO:0048318) |
| 0.1 |
0.2 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.1 |
1.7 |
GO:2000300 |
regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 |
0.9 |
GO:1904714 |
regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 |
0.2 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 |
0.7 |
GO:0001946 |
lymphangiogenesis(GO:0001946) |
| 0.1 |
0.2 |
GO:1903215 |
negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 |
0.2 |
GO:0038163 |
thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 |
0.7 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.1 |
0.7 |
GO:0035279 |
mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 |
0.9 |
GO:0034776 |
response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.1 |
0.1 |
GO:0015960 |
diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.1 |
0.3 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 |
1.9 |
GO:0048025 |
negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 |
0.3 |
GO:0044565 |
dendritic cell proliferation(GO:0044565) |
| 0.1 |
0.4 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 |
0.8 |
GO:0045956 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 |
0.3 |
GO:0048865 |
ganglion mother cell fate determination(GO:0007402) stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.1 |
0.5 |
GO:1903546 |
protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 |
0.1 |
GO:0035672 |
oligopeptide transmembrane transport(GO:0035672) |
| 0.1 |
0.2 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 |
0.2 |
GO:0070124 |
mitochondrial translational initiation(GO:0070124) |
| 0.1 |
0.1 |
GO:0035564 |
regulation of kidney size(GO:0035564) |
| 0.1 |
0.2 |
GO:0000711 |
meiotic DNA repair synthesis(GO:0000711) |
| 0.1 |
0.2 |
GO:1901656 |
glycoside transport(GO:1901656) |
| 0.1 |
0.1 |
GO:0034443 |
regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.1 |
0.1 |
GO:0060179 |
male mating behavior(GO:0060179) |
| 0.1 |
0.1 |
GO:0007056 |
spindle assembly involved in female meiosis(GO:0007056) |
| 0.1 |
0.5 |
GO:2001199 |
negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.1 |
0.2 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.1 |
0.2 |
GO:0097070 |
ductus arteriosus closure(GO:0097070) |
| 0.1 |
0.4 |
GO:0002667 |
lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.1 |
0.2 |
GO:1902460 |
regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 |
0.4 |
GO:0046325 |
negative regulation of glucose import(GO:0046325) |
| 0.1 |
0.3 |
GO:2000065 |
negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.1 |
0.3 |
GO:1903243 |
negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 |
0.1 |
GO:0045869 |
negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.1 |
0.1 |
GO:0086100 |
endothelin receptor signaling pathway(GO:0086100) |
| 0.1 |
0.1 |
GO:2000819 |
regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 |
0.1 |
GO:0021553 |
olfactory nerve development(GO:0021553) |
| 0.1 |
0.5 |
GO:0071313 |
cellular response to caffeine(GO:0071313) |
| 0.1 |
0.5 |
GO:0035020 |
regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 |
0.2 |
GO:0021612 |
facial nerve structural organization(GO:0021612) |
| 0.1 |
0.7 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.1 |
0.4 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.1 |
0.2 |
GO:0036289 |
peptidyl-serine autophosphorylation(GO:0036289) |
| 0.1 |
2.7 |
GO:0050690 |
regulation of defense response to virus by virus(GO:0050690) |
| 0.1 |
0.3 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 |
1.3 |
GO:2001222 |
regulation of neuron migration(GO:2001222) |
| 0.1 |
0.4 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 |
0.2 |
GO:0001188 |
RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.1 |
0.3 |
GO:1903963 |
arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 |
0.1 |
GO:0045198 |
establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 |
0.1 |
GO:0097106 |
postsynaptic density organization(GO:0097106) |
| 0.1 |
0.3 |
GO:0014063 |
regulation of serotonin secretion(GO:0014062) negative regulation of serotonin secretion(GO:0014063) |
| 0.1 |
0.3 |
GO:0003170 |
heart valve development(GO:0003170) |
| 0.1 |
0.1 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 |
0.1 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 |
0.1 |
GO:0033129 |
positive regulation of histone phosphorylation(GO:0033129) histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 |
0.1 |
GO:0045978 |
negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.1 |
0.2 |
GO:0016185 |
synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 |
0.6 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.1 |
0.4 |
GO:0033211 |
adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 |
1.0 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.1 |
0.6 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.1 |
0.2 |
GO:1903575 |
cornified envelope assembly(GO:1903575) |
| 0.1 |
0.2 |
GO:0075044 |
autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 |
0.5 |
GO:0060316 |
positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 |
0.3 |
GO:0042631 |
cellular response to water deprivation(GO:0042631) |
| 0.1 |
0.6 |
GO:0015939 |
pantothenate metabolic process(GO:0015939) |
| 0.1 |
0.3 |
GO:0042997 |
negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 |
0.3 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 |
1.0 |
GO:0090140 |
regulation of mitochondrial fission(GO:0090140) |
| 0.1 |
0.1 |
GO:0002581 |
negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 |
0.2 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 |
0.5 |
GO:0000186 |
activation of MAPKK activity(GO:0000186) |
| 0.1 |
0.1 |
GO:0051659 |
maintenance of mitochondrion location(GO:0051659) |
| 0.1 |
0.2 |
GO:1903412 |
response to bile acid(GO:1903412) |
| 0.1 |
0.2 |
GO:0051352 |
negative regulation of ligase activity(GO:0051352) |
| 0.1 |
0.2 |
GO:0014736 |
regulation of muscle atrophy(GO:0014735) negative regulation of muscle atrophy(GO:0014736) response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 |
0.1 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.1 |
0.3 |
GO:0030728 |
ovulation(GO:0030728) |
| 0.1 |
0.1 |
GO:0035669 |
TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.1 |
0.3 |
GO:0006522 |
alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.1 |
0.2 |
GO:0051685 |
endoplasmic reticulum localization(GO:0051643) maintenance of ER location(GO:0051685) |
| 0.1 |
0.4 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 |
0.1 |
GO:0010800 |
positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 |
0.1 |
GO:0021785 |
branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 |
0.8 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 |
0.2 |
GO:0010793 |
regulation of mRNA export from nucleus(GO:0010793) |
| 0.1 |
0.8 |
GO:0019934 |
cGMP-mediated signaling(GO:0019934) |
| 0.1 |
1.0 |
GO:0034199 |
activation of protein kinase A activity(GO:0034199) |
| 0.1 |
1.4 |
GO:1903846 |
positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 |
0.8 |
GO:0019371 |
cyclooxygenase pathway(GO:0019371) |
| 0.1 |
0.1 |
GO:1902548 |
negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.1 |
0.1 |
GO:0048341 |
paraxial mesoderm formation(GO:0048341) |
| 0.1 |
0.5 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.1 |
0.2 |
GO:0051136 |
regulation of NK T cell differentiation(GO:0051136) |
| 0.1 |
0.1 |
GO:1901205 |
negative regulation of adrenergic receptor signaling pathway(GO:0071878) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) negative regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901205) |
| 0.1 |
0.4 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) |
| 0.1 |
0.2 |
GO:1990502 |
dense core granule maturation(GO:1990502) |
| 0.1 |
0.2 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.1 |
2.2 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.1 |
0.8 |
GO:2001197 |
regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 |
0.3 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.1 |
0.4 |
GO:0070315 |
G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 |
0.3 |
GO:0038042 |
dimeric G-protein coupled receptor signaling pathway(GO:0038042) |
| 0.1 |
1.6 |
GO:0032469 |
endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 |
0.2 |
GO:0070212 |
protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 |
0.2 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 |
0.3 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 |
0.2 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 0.1 |
0.3 |
GO:0035810 |
positive regulation of urine volume(GO:0035810) |
| 0.1 |
0.1 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.1 |
0.2 |
GO:0001694 |
histamine biosynthetic process(GO:0001694) |
| 0.1 |
0.3 |
GO:0060841 |
venous blood vessel development(GO:0060841) |
| 0.1 |
0.1 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 |
0.4 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 |
0.1 |
GO:0006651 |
diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 |
0.2 |
GO:0090598 |
male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 |
0.4 |
GO:0035093 |
spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 |
0.2 |
GO:0061588 |
calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 |
0.3 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.1 |
0.3 |
GO:0065002 |
intracellular protein transmembrane transport(GO:0065002) |
| 0.1 |
0.5 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.1 |
0.1 |
GO:0090427 |
activation of meiosis(GO:0090427) |
| 0.1 |
0.3 |
GO:0001954 |
positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 |
0.1 |
GO:1902219 |
negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 |
0.3 |
GO:1900274 |
regulation of phospholipase C activity(GO:1900274) |
| 0.0 |
0.1 |
GO:0009786 |
regulation of asymmetric cell division(GO:0009786) |
| 0.0 |
0.1 |
GO:0042428 |
serotonin metabolic process(GO:0042428) |
| 0.0 |
0.5 |
GO:0045945 |
positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 |
0.1 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 |
0.6 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 |
0.1 |
GO:0038193 |
thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 |
0.2 |
GO:0006982 |
response to lipid hydroperoxide(GO:0006982) |
| 0.0 |
0.0 |
GO:1903542 |
negative regulation of exosomal secretion(GO:1903542) |
| 0.0 |
0.0 |
GO:1902913 |
positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.0 |
0.2 |
GO:0001306 |
age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 |
0.7 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.0 |
0.2 |
GO:0051673 |
membrane disruption in other organism(GO:0051673) |
| 0.0 |
0.7 |
GO:0036315 |
cellular response to sterol(GO:0036315) |
| 0.0 |
0.6 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.0 |
0.1 |
GO:0051409 |
response to nitrosative stress(GO:0051409) |
| 0.0 |
0.5 |
GO:0061087 |
positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 |
0.1 |
GO:0071947 |
protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 |
0.2 |
GO:1901300 |
positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.0 |
0.2 |
GO:0008588 |
release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 |
0.1 |
GO:0045994 |
positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 |
0.0 |
GO:0097676 |
histone H3-K36 dimethylation(GO:0097676) |
| 0.0 |
0.5 |
GO:0010968 |
regulation of microtubule nucleation(GO:0010968) |
| 0.0 |
0.1 |
GO:0086028 |
bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 |
0.1 |
GO:0006427 |
histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 |
0.2 |
GO:0070837 |
dehydroascorbic acid transport(GO:0070837) |
| 0.0 |
0.0 |
GO:0061469 |
regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 |
0.2 |
GO:0001878 |
response to yeast(GO:0001878) |
| 0.0 |
0.0 |
GO:0007127 |
meiosis I(GO:0007127) |
| 0.0 |
0.3 |
GO:0060693 |
regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 |
0.2 |
GO:0007512 |
adult heart development(GO:0007512) |
| 0.0 |
0.1 |
GO:0001541 |
ovarian follicle development(GO:0001541) |
| 0.0 |
0.4 |
GO:0046604 |
positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 |
1.2 |
GO:0006469 |
negative regulation of protein kinase activity(GO:0006469) |
| 0.0 |
0.0 |
GO:1903553 |
positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 |
0.1 |
GO:1900242 |
regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 |
0.2 |
GO:0003366 |
cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 |
0.1 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 |
0.2 |
GO:0002281 |
macrophage activation involved in immune response(GO:0002281) |
| 0.0 |
1.4 |
GO:0009435 |
NAD biosynthetic process(GO:0009435) |
| 0.0 |
2.6 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.1 |
GO:0019087 |
transformation of host cell by virus(GO:0019087) |
| 0.0 |
0.3 |
GO:0071243 |
cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 |
0.0 |
GO:1902463 |
protein localization to cell leading edge(GO:1902463) |
| 0.0 |
0.8 |
GO:0010765 |
positive regulation of sodium ion transport(GO:0010765) |
| 0.0 |
0.1 |
GO:0014887 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 |
0.3 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 |
0.2 |
GO:0003433 |
chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 |
0.2 |
GO:0072025 |
distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.0 |
1.9 |
GO:0097061 |
dendritic spine organization(GO:0097061) |
| 0.0 |
0.3 |
GO:0040032 |
parathyroid hormone secretion(GO:0035898) post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 |
0.3 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.0 |
0.3 |
GO:0010875 |
positive regulation of cholesterol efflux(GO:0010875) |
| 0.0 |
0.4 |
GO:1902902 |
negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 |
0.2 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.0 |
0.1 |
GO:1905154 |
negative regulation of membrane invagination(GO:1905154) |
| 0.0 |
0.7 |
GO:0042340 |
keratan sulfate catabolic process(GO:0042340) |
| 0.0 |
0.0 |
GO:0051459 |
regulation of corticotropin secretion(GO:0051459) |
| 0.0 |
0.1 |
GO:0038188 |
cholecystokinin signaling pathway(GO:0038188) |
| 0.0 |
0.9 |
GO:0070306 |
lens fiber cell differentiation(GO:0070306) |
| 0.0 |
0.3 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.0 |
0.1 |
GO:1900736 |
regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.0 |
0.5 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.0 |
GO:0010749 |
regulation of nitric oxide mediated signal transduction(GO:0010749) |
| 0.0 |
0.3 |
GO:0015705 |
iodide transport(GO:0015705) |
| 0.0 |
0.2 |
GO:0002028 |
regulation of sodium ion transport(GO:0002028) |
| 0.0 |
0.3 |
GO:0031584 |
activation of phospholipase D activity(GO:0031584) |
| 0.0 |
0.4 |
GO:0033622 |
integrin activation(GO:0033622) |
| 0.0 |
0.1 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 |
0.7 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.0 |
GO:2000812 |
regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 |
0.6 |
GO:0046629 |
gamma-delta T cell activation(GO:0046629) |
| 0.0 |
0.0 |
GO:0070228 |
lymphocyte apoptotic process(GO:0070227) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.0 |
0.1 |
GO:1904808 |
regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.0 |
1.1 |
GO:0007257 |
activation of JUN kinase activity(GO:0007257) |
| 0.0 |
0.4 |
GO:0030539 |
male genitalia development(GO:0030539) |
| 0.0 |
0.0 |
GO:0090131 |
mesenchyme migration(GO:0090131) |
| 0.0 |
0.3 |
GO:0006572 |
tyrosine catabolic process(GO:0006572) |
| 0.0 |
0.0 |
GO:0002874 |
regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 |
0.4 |
GO:0002544 |
chronic inflammatory response(GO:0002544) |
| 0.0 |
0.2 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
0.1 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 |
0.4 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 |
0.2 |
GO:0002934 |
desmosome organization(GO:0002934) |
| 0.0 |
0.1 |
GO:1900409 |
positive regulation of cellular response to oxidative stress(GO:1900409) |
| 0.0 |
0.1 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.0 |
0.2 |
GO:0055064 |
chloride ion homeostasis(GO:0055064) |
| 0.0 |
0.0 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 |
0.2 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.0 |
0.2 |
GO:0043932 |
ossification involved in bone remodeling(GO:0043932) |
| 0.0 |
0.1 |
GO:0009138 |
pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 |
0.1 |
GO:0071550 |
death-inducing signaling complex assembly(GO:0071550) |
| 0.0 |
0.9 |
GO:0035767 |
endothelial cell chemotaxis(GO:0035767) |
| 0.0 |
0.8 |
GO:0030866 |
cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
0.5 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
0.9 |
GO:0035589 |
G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 |
0.4 |
GO:0015871 |
choline transport(GO:0015871) |
| 0.0 |
0.1 |
GO:1904562 |
phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 |
0.1 |
GO:0052553 |
induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 |
0.2 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.0 |
0.1 |
GO:0072718 |
response to cisplatin(GO:0072718) |
| 0.0 |
0.1 |
GO:0002763 |
positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 |
2.1 |
GO:0046580 |
negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 |
0.2 |
GO:0003085 |
negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.0 |
0.2 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.1 |
GO:0060823 |
canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 |
0.6 |
GO:0090110 |
cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 |
1.0 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 |
0.2 |
GO:0046108 |
uridine metabolic process(GO:0046108) |
| 0.0 |
0.3 |
GO:0048712 |
negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 |
0.1 |
GO:0019046 |
release from viral latency(GO:0019046) |
| 0.0 |
0.7 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.2 |
GO:0043950 |
positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 |
0.1 |
GO:1903336 |
negative regulation of vacuolar transport(GO:1903336) |
| 0.0 |
0.2 |
GO:0044070 |
regulation of anion transport(GO:0044070) |
| 0.0 |
0.4 |
GO:0009134 |
nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 |
0.2 |
GO:1904327 |
protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 |
0.2 |
GO:0035092 |
sperm chromatin condensation(GO:0035092) |
| 0.0 |
0.2 |
GO:0009822 |
alkaloid catabolic process(GO:0009822) |
| 0.0 |
0.4 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.3 |
GO:2000622 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 |
0.2 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 |
0.4 |
GO:0060088 |
auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 |
0.2 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 |
0.3 |
GO:0051156 |
glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 |
0.3 |
GO:0061072 |
iris morphogenesis(GO:0061072) |
| 0.0 |
0.0 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 |
0.1 |
GO:0051446 |
positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 |
0.1 |
GO:0060282 |
positive regulation of oocyte development(GO:0060282) |
| 0.0 |
0.1 |
GO:0099612 |
protein localization to axon(GO:0099612) |
| 0.0 |
0.0 |
GO:0035995 |
detection of muscle stretch(GO:0035995) |
| 0.0 |
0.6 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.7 |
GO:0042994 |
cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 |
0.1 |
GO:0006478 |
peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 |
0.1 |
GO:0090675 |
intermicrovillar adhesion(GO:0090675) |
| 0.0 |
0.2 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 |
0.3 |
GO:0060770 |
negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 |
0.6 |
GO:0043651 |
linoleic acid metabolic process(GO:0043651) |
| 0.0 |
0.8 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.3 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.0 |
GO:0002188 |
translation reinitiation(GO:0002188) |
| 0.0 |
0.1 |
GO:0060336 |
negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) cellular response to vitamin E(GO:0071306) response to metformin(GO:1901558) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 |
0.4 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.1 |
GO:0035878 |
nail development(GO:0035878) |
| 0.0 |
0.1 |
GO:0035641 |
locomotory exploration behavior(GO:0035641) |
| 0.0 |
0.5 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.0 |
0.2 |
GO:0035105 |
sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 |
0.8 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.3 |
GO:0015732 |
prostaglandin transport(GO:0015732) |
| 0.0 |
0.1 |
GO:0072553 |
terminal button organization(GO:0072553) |
| 0.0 |
0.7 |
GO:0048854 |
brain morphogenesis(GO:0048854) |
| 0.0 |
0.1 |
GO:0071418 |
cellular response to amine stimulus(GO:0071418) |
| 0.0 |
0.1 |
GO:1902230 |
negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 |
1.0 |
GO:0003254 |
regulation of membrane depolarization(GO:0003254) |
| 0.0 |
0.1 |
GO:1901069 |
guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 |
0.4 |
GO:0060046 |
regulation of acrosome reaction(GO:0060046) |
| 0.0 |
0.7 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 |
0.5 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.0 |
0.1 |
GO:0090521 |
glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 |
0.3 |
GO:0045649 |
regulation of macrophage differentiation(GO:0045649) |
| 0.0 |
0.3 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.0 |
0.7 |
GO:0030252 |
growth hormone secretion(GO:0030252) |
| 0.0 |
0.1 |
GO:0001777 |
T cell homeostatic proliferation(GO:0001777) |
| 0.0 |
0.1 |
GO:1990737 |
response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 |
0.2 |
GO:0015840 |
urea transport(GO:0015840) |
| 0.0 |
0.1 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.0 |
0.3 |
GO:0098787 |
mRNA cleavage involved in mRNA processing(GO:0098787) |
| 0.0 |
0.2 |
GO:0044539 |
long-chain fatty acid import(GO:0044539) |
| 0.0 |
0.1 |
GO:0035999 |
tetrahydrofolate interconversion(GO:0035999) |
| 0.0 |
0.1 |
GO:0048537 |
mucosal-associated lymphoid tissue development(GO:0048537) Peyer's patch development(GO:0048541) Peyer's patch morphogenesis(GO:0061146) |
| 0.0 |
0.1 |
GO:0000117 |
regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 |
0.2 |
GO:0050777 |
negative regulation of immune response(GO:0050777) |
| 0.0 |
0.2 |
GO:1901503 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 |
0.2 |
GO:0019556 |
histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 |
0.3 |
GO:0002414 |
immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 |
0.1 |
GO:0061303 |
cornea development in camera-type eye(GO:0061303) |
| 0.0 |
0.2 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 |
0.2 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.0 |
0.2 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.0 |
0.3 |
GO:0021854 |
hypothalamus development(GO:0021854) |
| 0.0 |
0.3 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 |
0.0 |
GO:0071321 |
cellular response to cGMP(GO:0071321) |
| 0.0 |
0.3 |
GO:0042167 |
porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 |
0.2 |
GO:0032288 |
myelin assembly(GO:0032288) |
| 0.0 |
0.1 |
GO:1903597 |
negative regulation of gap junction assembly(GO:1903597) positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.0 |
0.3 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.1 |
GO:0050928 |
negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 |
0.3 |
GO:0032510 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 |
1.9 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.0 |
0.0 |
GO:0046351 |
disaccharide biosynthetic process(GO:0046351) |
| 0.0 |
0.2 |
GO:0000389 |
mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 |
0.2 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.1 |
GO:0060697 |
positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 |
0.1 |
GO:2001300 |
lipoxin metabolic process(GO:2001300) |
| 0.0 |
0.2 |
GO:0009071 |
serine family amino acid catabolic process(GO:0009071) |
| 0.0 |
0.2 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.0 |
0.0 |
GO:0050666 |
regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.0 |
0.0 |
GO:0032757 |
positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 |
0.1 |
GO:0035879 |
plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.8 |
GO:0045879 |
negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 |
0.5 |
GO:0060330 |
regulation of response to interferon-gamma(GO:0060330) |
| 0.0 |
0.6 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.0 |
0.1 |
GO:0002643 |
regulation of tolerance induction(GO:0002643) |
| 0.0 |
0.3 |
GO:0042753 |
positive regulation of circadian rhythm(GO:0042753) |
| 0.0 |
0.2 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.0 |
0.1 |
GO:0009609 |
response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.0 |
0.2 |
GO:0010523 |
negative regulation of calcium ion transport into cytosol(GO:0010523) |
| 0.0 |
0.1 |
GO:0033031 |
positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 |
0.2 |
GO:0035617 |
stress granule disassembly(GO:0035617) |
| 0.0 |
0.3 |
GO:0045776 |
negative regulation of blood pressure(GO:0045776) |
| 0.0 |
0.1 |
GO:0046833 |
positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 |
0.1 |
GO:0060054 |
positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 |
0.3 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 |
0.1 |
GO:1903351 |
response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.0 |
0.2 |
GO:0010248 |
establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 |
0.1 |
GO:0034971 |
histone H3-R17 methylation(GO:0034971) |
| 0.0 |
0.6 |
GO:0034755 |
iron ion transmembrane transport(GO:0034755) |
| 0.0 |
0.2 |
GO:0019470 |
4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 |
0.2 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
0.1 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.0 |
0.1 |
GO:0071442 |
regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 |
0.0 |
GO:1990168 |
protein K33-linked deubiquitination(GO:1990168) |
| 0.0 |
0.1 |
GO:0055069 |
zinc ion homeostasis(GO:0055069) |
| 0.0 |
0.3 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.1 |
GO:0072268 |
pattern specification involved in metanephros development(GO:0072268) |
| 0.0 |
0.1 |
GO:0035518 |
histone H2A monoubiquitination(GO:0035518) |
| 0.0 |
0.2 |
GO:1904706 |
negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 |
0.0 |
GO:0043335 |
protein unfolding(GO:0043335) |
| 0.0 |
0.8 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.0 |
0.1 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.0 |
0.6 |
GO:0090004 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 |
0.2 |
GO:0002329 |
pre-B cell differentiation(GO:0002329) |
| 0.0 |
0.2 |
GO:0019896 |
axonal transport of mitochondrion(GO:0019896) |
| 0.0 |
0.1 |
GO:0051001 |
negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 |
0.1 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 |
0.2 |
GO:0003091 |
renal water homeostasis(GO:0003091) |
| 0.0 |
0.3 |
GO:0043278 |
response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 |
0.4 |
GO:1901750 |
leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 |
0.3 |
GO:0010831 |
positive regulation of myotube differentiation(GO:0010831) |
| 0.0 |
0.4 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.0 |
0.3 |
GO:0016264 |
gap junction assembly(GO:0016264) |
| 0.0 |
0.7 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.8 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.1 |
GO:0007343 |
egg activation(GO:0007343) |
| 0.0 |
0.1 |
GO:0043456 |
regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 |
0.2 |
GO:0090261 |
positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 |
0.1 |
GO:0051932 |
synaptic transmission, GABAergic(GO:0051932) |
| 0.0 |
0.2 |
GO:0045668 |
negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 |
0.1 |
GO:1904352 |
positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 |
0.1 |
GO:1901350 |
cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 |
0.0 |
GO:0030432 |
peristalsis(GO:0030432) |
| 0.0 |
0.1 |
GO:0000212 |
meiotic spindle organization(GO:0000212) |
| 0.0 |
0.1 |
GO:1904647 |
response to rotenone(GO:1904647) |
| 0.0 |
0.3 |
GO:0007635 |
chemosensory behavior(GO:0007635) |
| 0.0 |
1.6 |
GO:0070098 |
chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 |
0.1 |
GO:0030431 |
sleep(GO:0030431) |
| 0.0 |
0.0 |
GO:0050654 |
chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.0 |
0.1 |
GO:0070365 |
hepatocyte differentiation(GO:0070365) |
| 0.0 |
1.4 |
GO:0035036 |
sperm-egg recognition(GO:0035036) |
| 0.0 |
0.3 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.0 |
0.1 |
GO:0071955 |
recycling endosome to Golgi transport(GO:0071955) |
| 0.0 |
0.2 |
GO:0000414 |
regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 |
0.8 |
GO:0048008 |
platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 |
0.4 |
GO:0018904 |
ether metabolic process(GO:0018904) |
| 0.0 |
0.1 |
GO:0051354 |
negative regulation of oxidoreductase activity(GO:0051354) |
| 0.0 |
0.1 |
GO:0045617 |
negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 |
0.4 |
GO:0001514 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.2 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 |
1.2 |
GO:0006939 |
smooth muscle contraction(GO:0006939) |
| 0.0 |
0.3 |
GO:0046626 |
regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 |
1.2 |
GO:0018279 |
protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 |
0.8 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.1 |
GO:0002118 |
aggressive behavior(GO:0002118) |
| 0.0 |
0.2 |
GO:0045187 |
regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 |
0.1 |
GO:0046440 |
L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 |
0.5 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 |
0.0 |
GO:0006579 |
amino-acid betaine catabolic process(GO:0006579) |
| 0.0 |
0.2 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.0 |
0.2 |
GO:0006574 |
valine catabolic process(GO:0006574) |
| 0.0 |
0.4 |
GO:0046051 |
UTP metabolic process(GO:0046051) |
| 0.0 |
0.1 |
GO:0034145 |
positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 |
0.2 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 |
0.2 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.0 |
0.5 |
GO:0042501 |
serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 |
0.3 |
GO:2000392 |
regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 |
0.1 |
GO:0032536 |
regulation of cell projection size(GO:0032536) |
| 0.0 |
0.3 |
GO:0045109 |
intermediate filament organization(GO:0045109) |
| 0.0 |
0.1 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.1 |
GO:0070585 |
protein localization to mitochondrion(GO:0070585) |
| 0.0 |
0.2 |
GO:0032693 |
negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 |
0.2 |
GO:0098704 |
fructose transport(GO:0015755) fructose import(GO:0032445) cellular response to fructose stimulus(GO:0071332) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 |
0.2 |
GO:2000535 |
regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 |
0.0 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 |
0.9 |
GO:0006739 |
NADP metabolic process(GO:0006739) |
| 0.0 |
0.4 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.0 |
0.1 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.0 |
0.2 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.0 |
0.1 |
GO:0006884 |
cell volume homeostasis(GO:0006884) |
| 0.0 |
0.6 |
GO:0050819 |
negative regulation of coagulation(GO:0050819) |
| 0.0 |
0.1 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 |
0.2 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
0.0 |
GO:0042214 |
terpene metabolic process(GO:0042214) |
| 0.0 |
0.2 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.0 |
0.4 |
GO:0006625 |
protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 |
0.0 |
GO:2000110 |
negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 |
0.5 |
GO:0036150 |
phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 |
0.3 |
GO:0060732 |
positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 |
0.1 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 |
0.1 |
GO:0010533 |
regulation of activation of Janus kinase activity(GO:0010533) positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 |
0.0 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.0 |
0.3 |
GO:0006307 |
DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 |
0.1 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 |
0.1 |
GO:0033689 |
negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 |
0.1 |
GO:0001974 |
blood vessel remodeling(GO:0001974) |
| 0.0 |
0.6 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 |
0.1 |
GO:1902410 |
mitotic cytokinetic process(GO:1902410) |
| 0.0 |
0.3 |
GO:1900116 |
extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 |
0.1 |
GO:0010735 |
positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 |
0.2 |
GO:0097396 |
response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 |
0.1 |
GO:0061113 |
pancreas morphogenesis(GO:0061113) |
| 0.0 |
0.1 |
GO:0010666 |
positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 |
0.1 |
GO:0032495 |
response to muramyl dipeptide(GO:0032495) |
| 0.0 |
0.0 |
GO:0071504 |
cellular response to heparin(GO:0071504) |
| 0.0 |
0.2 |
GO:0006089 |
lactate metabolic process(GO:0006089) |
| 0.0 |
0.2 |
GO:0097178 |
ruffle assembly(GO:0097178) |
| 0.0 |
0.7 |
GO:0007189 |
adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 |
0.2 |
GO:0031086 |
nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) |
| 0.0 |
0.3 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 |
0.4 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.1 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 |
0.1 |
GO:0071163 |
DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 |
0.1 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 |
0.2 |
GO:1901838 |
positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 |
0.1 |
GO:0021615 |
glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 |
0.1 |
GO:0042435 |
indole-containing compound biosynthetic process(GO:0042435) indolalkylamine biosynthetic process(GO:0046219) |
| 0.0 |
0.1 |
GO:0043383 |
negative T cell selection(GO:0043383) |
| 0.0 |
0.3 |
GO:0070933 |
histone H4 deacetylation(GO:0070933) |
| 0.0 |
0.1 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.1 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 |
0.1 |
GO:0044130 |
negative regulation of growth of symbiont in host(GO:0044130) |
| 0.0 |
0.1 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.0 |
0.0 |
GO:1901843 |
positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 |
0.1 |
GO:0030168 |
platelet activation(GO:0030168) |
| 0.0 |
0.3 |
GO:0030521 |
androgen receptor signaling pathway(GO:0030521) |
| 0.0 |
0.1 |
GO:0098700 |
neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 |
0.1 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.0 |
0.1 |
GO:0038155 |
interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 |
0.0 |
GO:2000609 |
regulation of thyroid hormone generation(GO:2000609) |
| 0.0 |
0.1 |
GO:0044778 |
meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 |
0.1 |
GO:0045876 |
positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 |
0.9 |
GO:0038083 |
peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 |
0.1 |
GO:0016240 |
autophagosome docking(GO:0016240) |
| 0.0 |
0.0 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.0 |
0.1 |
GO:0010836 |
negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 |
0.1 |
GO:0090136 |
epithelial cell-cell adhesion(GO:0090136) |
| 0.0 |
0.4 |
GO:0007340 |
acrosome reaction(GO:0007340) |
| 0.0 |
0.1 |
GO:0007320 |
insemination(GO:0007320) |
| 0.0 |
0.0 |
GO:0043931 |
ossification involved in bone maturation(GO:0043931) |
| 0.0 |
0.1 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 |
0.0 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.0 |
GO:0090235 |
regulation of metaphase plate congression(GO:0090235) |
| 0.0 |
0.2 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.0 |
0.1 |
GO:0061737 |
leukotriene signaling pathway(GO:0061737) |
| 0.0 |
0.2 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 |
0.4 |
GO:0061718 |
NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 |
0.3 |
GO:0042059 |
negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.0 |
0.1 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 |
0.0 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.0 |
0.1 |
GO:0008218 |
bioluminescence(GO:0008218) |
| 0.0 |
0.0 |
GO:0032494 |
response to peptidoglycan(GO:0032494) |
| 0.0 |
0.3 |
GO:0019388 |
galactose catabolic process(GO:0019388) |
| 0.0 |
0.2 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 |
0.1 |
GO:0034384 |
high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 |
0.3 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.0 |
0.1 |
GO:0045656 |
negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 |
0.1 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 |
0.1 |
GO:0035633 |
maintenance of blood-brain barrier(GO:0035633) |
| 0.0 |
0.1 |
GO:0048549 |
positive regulation of pinocytosis(GO:0048549) |
| 0.0 |
0.2 |
GO:0010885 |
regulation of cholesterol storage(GO:0010885) |
| 0.0 |
0.1 |
GO:0090043 |
tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.0 |
0.2 |
GO:0046618 |
drug export(GO:0046618) |
| 0.0 |
0.1 |
GO:0070586 |
cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.0 |
0.1 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.0 |
0.0 |
GO:0010713 |
negative regulation of collagen metabolic process(GO:0010713) negative regulation of multicellular organismal metabolic process(GO:0044252) |
| 0.0 |
0.1 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 |
0.0 |
GO:0006544 |
glycine metabolic process(GO:0006544) |
| 0.0 |
0.1 |
GO:0048169 |
regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 |
0.1 |
GO:0015909 |
long-chain fatty acid transport(GO:0015909) |
| 0.0 |
0.3 |
GO:0043001 |
Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 |
0.1 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 |
0.1 |
GO:0032354 |
response to follicle-stimulating hormone(GO:0032354) |
| 0.0 |
0.1 |
GO:0090186 |
regulation of pancreatic juice secretion(GO:0090186) |
| 0.0 |
0.1 |
GO:0019919 |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) peptidyl-arginine N-methylation(GO:0035246) peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.0 |
0.1 |
GO:0030047 |
actin modification(GO:0030047) |
| 0.0 |
0.1 |
GO:0000706 |
meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 |
0.1 |
GO:0042908 |
xenobiotic transport(GO:0042908) |
| 0.0 |
0.1 |
GO:0033490 |
cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 |
0.0 |
GO:0070858 |
negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 |
0.2 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
1.1 |
GO:0036498 |
IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 |
1.0 |
GO:1903779 |
regulation of cardiac conduction(GO:1903779) |
| 0.0 |
0.1 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.0 |
GO:0015695 |
organic cation transport(GO:0015695) |
| 0.0 |
0.1 |
GO:0035562 |
negative regulation of chromatin binding(GO:0035562) |
| 0.0 |
0.1 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 |
0.5 |
GO:0007173 |
epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 |
0.1 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.0 |
0.0 |
GO:0002666 |
positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 |
0.1 |
GO:0016098 |
monoterpenoid metabolic process(GO:0016098) |
| 0.0 |
0.1 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.0 |
0.1 |
GO:0050951 |
sensory perception of temperature stimulus(GO:0050951) |
| 0.0 |
0.1 |
GO:0071896 |
protein localization to adherens junction(GO:0071896) |
| 0.0 |
0.1 |
GO:0032596 |
protein transport into membrane raft(GO:0032596) |
| 0.0 |
0.2 |
GO:0086010 |
membrane depolarization during action potential(GO:0086010) |
| 0.0 |
0.1 |
GO:0010889 |
regulation of sequestering of triglyceride(GO:0010889) sequestering of triglyceride(GO:0030730) |
| 0.0 |
0.0 |
GO:1901207 |
regulation of heart looping(GO:1901207) |
| 0.0 |
0.0 |
GO:1902908 |
regulation of melanosome transport(GO:1902908) |
| 0.0 |
0.5 |
GO:1903959 |
regulation of anion transmembrane transport(GO:1903959) |
| 0.0 |
0.0 |
GO:0060012 |
synaptic transmission, glycinergic(GO:0060012) |
| 0.0 |
0.1 |
GO:0001556 |
oocyte maturation(GO:0001556) |
| 0.0 |
0.1 |
GO:0032824 |
natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.0 |
0.0 |
GO:1901894 |
regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 |
0.2 |
GO:0050982 |
detection of mechanical stimulus(GO:0050982) |
| 0.0 |
0.3 |
GO:0043029 |
T cell homeostasis(GO:0043029) |
| 0.0 |
0.1 |
GO:0098703 |
calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 |
0.0 |
GO:0001767 |
establishment of lymphocyte polarity(GO:0001767) |
| 0.0 |
0.4 |
GO:0045589 |
regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 |
0.1 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 |
0.0 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.0 |
0.1 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 |
0.1 |
GO:0007039 |
protein catabolic process in the vacuole(GO:0007039) |
| 0.0 |
0.1 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.0 |
0.2 |
GO:0050829 |
defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 |
0.1 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.0 |
0.1 |
GO:2000291 |
regulation of myoblast proliferation(GO:2000291) |
| 0.0 |
0.0 |
GO:0002572 |
pro-T cell differentiation(GO:0002572) |
| 0.0 |
0.1 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 |
0.0 |
GO:0043117 |
positive regulation of vascular permeability(GO:0043117) |
| 0.0 |
0.1 |
GO:0019062 |
virion attachment to host cell(GO:0019062) |
| 0.0 |
0.1 |
GO:0046719 |
regulation by virus of viral protein levels in host cell(GO:0046719) |
| 0.0 |
0.0 |
GO:0035359 |
negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 |
0.0 |
GO:0050849 |
negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 |
0.2 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.0 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.0 |
0.2 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
0.0 |
GO:0042754 |
negative regulation of circadian rhythm(GO:0042754) |
| 0.0 |
0.1 |
GO:0033561 |
regulation of water loss via skin(GO:0033561) |
| 0.0 |
2.3 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 |
0.1 |
GO:0032418 |
lysosome localization(GO:0032418) |
| 0.0 |
0.2 |
GO:0006309 |
apoptotic DNA fragmentation(GO:0006309) |
| 0.0 |
0.1 |
GO:0002097 |
tRNA wobble base modification(GO:0002097) |
| 0.0 |
0.1 |
GO:0046967 |
cytosol to ER transport(GO:0046967) |
| 0.0 |
0.0 |
GO:0007140 |
male meiosis(GO:0007140) |
| 0.0 |
0.1 |
GO:0034382 |
chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 |
0.7 |
GO:0050830 |
defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 |
1.1 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.8 |
GO:0006910 |
phagocytosis, recognition(GO:0006910) |
| 0.0 |
0.6 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.4 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 |
0.0 |
GO:0005997 |
xylulose metabolic process(GO:0005997) |
| 0.0 |
0.0 |
GO:1903674 |
regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 |
0.3 |
GO:0009303 |
rRNA transcription(GO:0009303) |
| 0.0 |
0.1 |
GO:0032098 |
regulation of appetite(GO:0032098) |
| 0.0 |
0.8 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.0 |
GO:2000825 |
positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 |
0.1 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 |
0.0 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.0 |
0.1 |
GO:0006706 |
steroid catabolic process(GO:0006706) |
| 0.0 |
0.5 |
GO:0043928 |
exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 |
0.0 |
GO:0030889 |
negative regulation of B cell proliferation(GO:0030889) |
| 0.0 |
0.1 |
GO:0043031 |
negative regulation of macrophage activation(GO:0043031) |
| 0.0 |
0.2 |
GO:0007215 |
glutamate receptor signaling pathway(GO:0007215) |
| 0.0 |
0.1 |
GO:0007567 |
parturition(GO:0007567) |
| 0.0 |
0.5 |
GO:0006099 |
tricarboxylic acid cycle(GO:0006099) |
| 0.0 |
0.0 |
GO:0018194 |
N-terminal protein amino acid methylation(GO:0006480) N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.0 |
0.1 |
GO:0006435 |
threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 |
0.0 |
GO:0044340 |
canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.0 |
0.1 |
GO:0061298 |
retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 |
0.1 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.0 |
0.1 |
GO:0006538 |
glutamate catabolic process(GO:0006538) |
| 0.0 |
0.0 |
GO:1900451 |
positive regulation of glutamate receptor signaling pathway(GO:1900451) |
| 0.0 |
0.0 |
GO:0006987 |
activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 |
0.1 |
GO:1902775 |
mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 |
0.1 |
GO:0060481 |
lobar bronchus epithelium development(GO:0060481) lobar bronchus development(GO:0060482) |
| 0.0 |
0.1 |
GO:0000720 |
pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 |
0.1 |
GO:0033227 |
dsRNA transport(GO:0033227) |
| 0.0 |
0.1 |
GO:0006558 |
L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 |
0.2 |
GO:0046320 |
regulation of fatty acid oxidation(GO:0046320) |
| 0.0 |
0.0 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 |
0.1 |
GO:0006189 |
'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 |
0.1 |
GO:0015942 |
formate metabolic process(GO:0015942) |
| 0.0 |
1.3 |
GO:0070268 |
cornification(GO:0070268) |
| 0.0 |
0.1 |
GO:1903027 |
regulation of opsonization(GO:1903027) |
| 0.0 |
0.1 |
GO:0071394 |
cellular response to testosterone stimulus(GO:0071394) |
| 0.0 |
0.1 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.0 |
0.0 |
GO:0032964 |
collagen biosynthetic process(GO:0032964) |
| 0.0 |
0.0 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 |
0.1 |
GO:0070301 |
cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 |
0.3 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.0 |
0.1 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
0.0 |
GO:0071460 |
cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 |
0.2 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.0 |
0.0 |
GO:0007028 |
cytoplasm organization(GO:0007028) |
| 0.0 |
0.0 |
GO:0071774 |
response to fibroblast growth factor(GO:0071774) |
| 0.0 |
0.1 |
GO:1901724 |
positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.0 |
0.0 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.0 |
0.1 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.0 |
GO:0046462 |
monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 |
0.0 |
GO:0021849 |
neuroblast division in subventricular zone(GO:0021849) |
| 0.0 |
0.0 |
GO:0070407 |
oxidation-dependent protein catabolic process(GO:0070407) |
| 0.0 |
0.0 |
GO:0071469 |
cellular response to alkaline pH(GO:0071469) |
| 0.0 |
0.0 |
GO:0097237 |
cellular response to toxic substance(GO:0097237) |
| 0.0 |
0.2 |
GO:0019730 |
antimicrobial humoral response(GO:0019730) |
| 0.0 |
0.2 |
GO:1901222 |
regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 |
0.1 |
GO:0070849 |
response to epidermal growth factor(GO:0070849) |
| 0.0 |
0.0 |
GO:0007172 |
signal complex assembly(GO:0007172) |
| 0.0 |
0.0 |
GO:0032438 |
melanosome organization(GO:0032438) |
| 0.0 |
0.0 |
GO:2000696 |
regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 |
0.1 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.0 |
0.0 |
GO:0043482 |
pigment accumulation(GO:0043476) cellular pigment accumulation(GO:0043482) |
| 0.0 |
0.1 |
GO:0007398 |
ectoderm development(GO:0007398) |
| 0.0 |
0.1 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.0 |
0.1 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 |
0.2 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 |
0.0 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.0 |
0.0 |
GO:2001140 |
regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.0 |
0.0 |
GO:0039535 |
regulation of RIG-I signaling pathway(GO:0039535) |
| 0.0 |
0.1 |
GO:0000050 |
urea cycle(GO:0000050) |
| 0.0 |
0.0 |
GO:0060167 |
regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.0 |
0.2 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.0 |
0.3 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 |
0.1 |
GO:0097411 |
hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 |
0.1 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.3 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.0 |
GO:0045065 |
cytotoxic T cell differentiation(GO:0045065) |
| 0.0 |
0.1 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 |
0.0 |
GO:0046122 |
purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 |
0.1 |
GO:0051341 |
regulation of oxidoreductase activity(GO:0051341) |
| 0.0 |
0.1 |
GO:0010960 |
magnesium ion homeostasis(GO:0010960) |
| 0.0 |
0.0 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 |
0.4 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.3 |
GO:0006027 |
glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 |
0.1 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 |
0.1 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.2 |
GO:0007566 |
embryo implantation(GO:0007566) |
| 0.0 |
0.0 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.0 |
0.1 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 |
0.1 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 |
0.1 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |