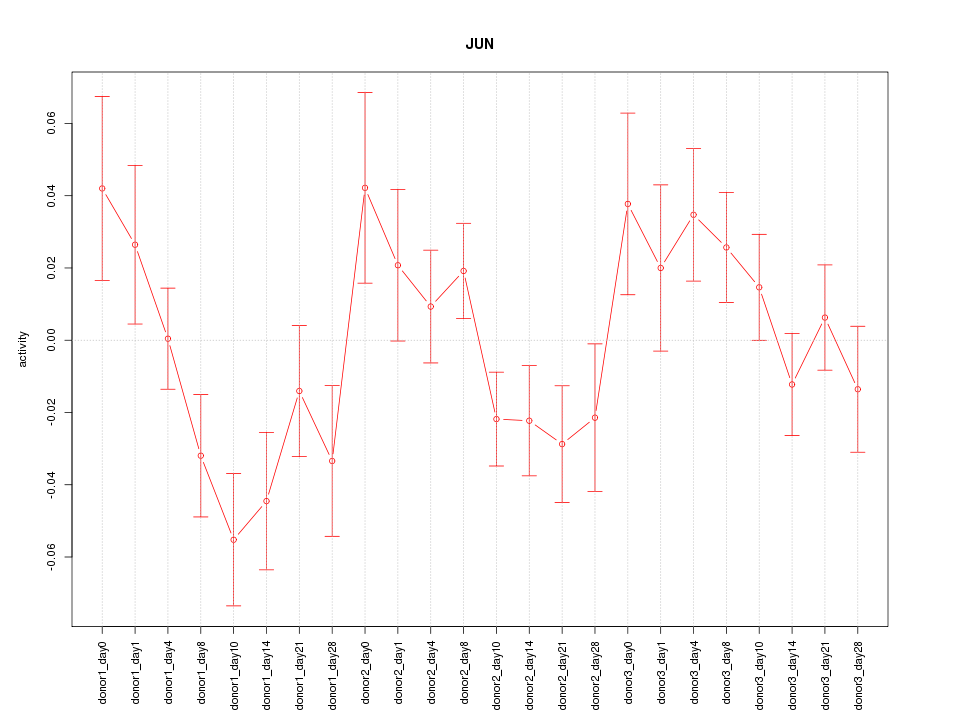
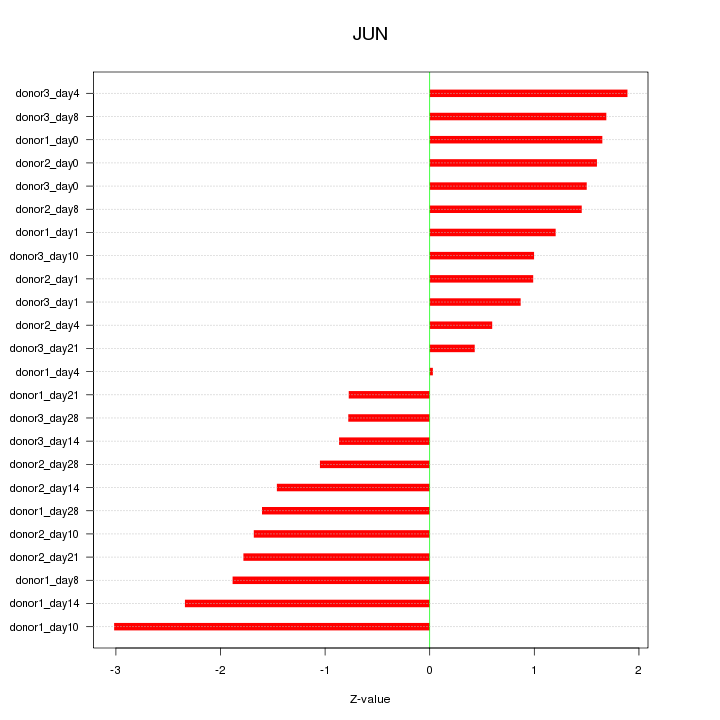
Motif ID: JUN
Z-value: 1.482
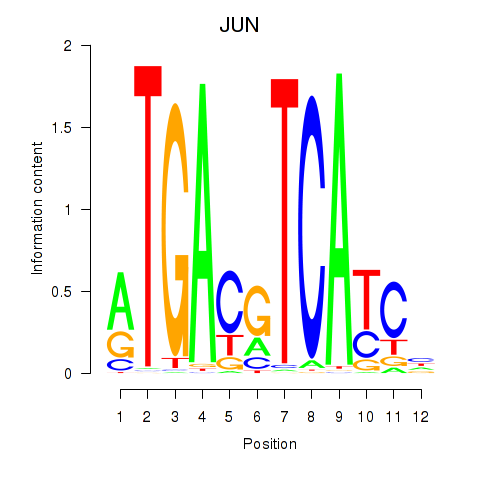
Transcription factors associated with JUN:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| JUN | ENSG00000177606.5 | JUN |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JUN | hg19_v2_chr1_-_59249732_59249785 | -0.41 | 4.7e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 2.2 | 6.6 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 1.3 | 3.8 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 1.3 | 7.5 | GO:0030035 | microspike assembly(GO:0030035) |
| 1.2 | 9.5 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 1.2 | 3.5 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 1.1 | 5.7 | GO:1904751 | positive regulation of telomeric DNA binding(GO:1904744) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.9 | 4.5 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.8 | 0.8 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.8 | 2.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.7 | 4.4 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.7 | 2.2 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.7 | 4.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.7 | 2.0 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.7 | 2.7 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.6 | 2.6 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.6 | 1.8 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.6 | 2.9 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.5 | 2.1 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.5 | 1.6 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.5 | 1.5 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.5 | 2.5 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.5 | 4.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.5 | 1.5 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.5 | 1.4 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.5 | 2.3 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.5 | 2.8 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.4 | 0.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.4 | 2.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.4 | 1.3 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.4 | 3.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.4 | 1.2 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.4 | 2.3 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.4 | 1.6 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.4 | 1.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.4 | 5.5 | GO:0000050 | urea cycle(GO:0000050) |
| 0.4 | 1.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.3 | 1.0 | GO:2000705 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.3 | 1.6 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.3 | 0.9 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.3 | 3.7 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 0.3 | GO:2000724 | positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.3 | 0.8 | GO:0052314 | terpene metabolic process(GO:0042214) phytoalexin metabolic process(GO:0052314) |
| 0.3 | 6.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 2.5 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 0.8 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.3 | 1.6 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.3 | 0.8 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.2 | 1.0 | GO:0006408 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.2 | 0.7 | GO:0060545 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) positive regulation of necroptotic process(GO:0060545) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.2 | 0.9 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) positive regulation of protein monoubiquitination(GO:1902527) |
| 0.2 | 0.9 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.2 | 2.3 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.2 | 0.9 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.2 | 0.7 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.2 | 3.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 0.6 | GO:0007343 | egg activation(GO:0007343) |
| 0.2 | 0.8 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 1.5 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 | 3.5 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 0.6 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 0.8 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.2 | 0.4 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.2 | 1.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 3.9 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.2 | 0.6 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.2 | 1.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.2 | 0.7 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.2 | 1.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.2 | 0.7 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 1.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.2 | 3.5 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.2 | 0.9 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.2 | 0.3 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.2 | 0.5 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.2 | 0.8 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.2 | 0.5 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.2 | 0.5 | GO:0021896 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.2 | 0.6 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.2 | 2.4 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.2 | 1.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 1.6 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.2 | 1.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.6 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.1 | 0.7 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.4 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.6 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.7 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) negative regulation of oocyte development(GO:0060283) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 7.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.4 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.1 | 0.5 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.5 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 1.5 | GO:0007135 | meiosis II(GO:0007135) |
| 0.1 | 2.8 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 3.4 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 0.4 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.6 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.4 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 1.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 1.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.8 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.6 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.6 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 1.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 1.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 2.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 0.5 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.4 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 1.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 2.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.5 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 0.3 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 0.6 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.1 | 0.8 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 2.4 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 1.0 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.1 | 0.3 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.1 | 2.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.5 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.3 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 | 0.9 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.3 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.4 | GO:0009953 | dorsal/ventral pattern formation(GO:0009953) |
| 0.1 | 0.4 | GO:0051272 | positive regulation of cellular component movement(GO:0051272) |
| 0.1 | 0.4 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 1.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.7 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.1 | 2.7 | GO:0000732 | strand displacement(GO:0000732) |
| 0.1 | 7.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 1.7 | GO:0030813 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.1 | 1.1 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.6 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.9 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 1.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 1.7 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.2 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 1.3 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 1.0 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.1 | 0.9 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.5 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 0.5 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.1 | 0.4 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.8 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.5 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.4 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 1.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.7 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.1 | 2.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.7 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.4 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.4 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 0.2 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.8 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 1.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.6 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 1.3 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.4 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.2 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 1.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.9 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 1.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.7 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.4 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.6 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.6 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.3 | GO:0071352 | cellular response to interleukin-2(GO:0071352) |
| 0.0 | 2.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0071224 | positive regulation of Schwann cell differentiation(GO:0014040) cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.5 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 3.3 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.3 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.3 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 1.4 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 1.4 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.8 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.4 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.8 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 1.0 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.6 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 1.4 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.3 | GO:0051493 | regulation of cytoskeleton organization(GO:0051493) |
| 0.0 | 0.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.2 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.5 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway(GO:0007169) |
| 0.0 | 0.4 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.1 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.0 | 1.3 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.6 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 | 0.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 9.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.2 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.8 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0051303 | establishment of chromosome localization(GO:0051303) |
| 0.0 | 2.0 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 1.6 | GO:0043525 | positive regulation of neuron apoptotic process(GO:0043525) |
| 0.0 | 1.6 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.5 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 1.2 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.7 | GO:0006468 | protein phosphorylation(GO:0006468) |
| 0.0 | 0.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.3 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 2.0 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 0.2 | GO:2001140 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.0 | 0.2 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.5 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.5 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 1.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.7 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.4 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.2 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.6 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.2 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.6 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.5 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 2.2 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.4 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.7 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.5 | GO:0043065 | positive regulation of apoptotic process(GO:0043065) |
| 0.0 | 0.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.0 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.6 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.7 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 1.2 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 1.8 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.3 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 2.7 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.3 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 1.1 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.5 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 0.4 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 2.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.5 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0031523 | Myb complex(GO:0031523) |
| 1.2 | 3.5 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.9 | 2.8 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.8 | 2.5 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.8 | 2.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.7 | 3.7 | GO:0044393 | microspike(GO:0044393) |
| 0.7 | 2.2 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.7 | 9.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.7 | 5.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.5 | 6.7 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.5 | 2.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.4 | 1.7 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.3 | 1.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.3 | 2.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 1.6 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.3 | 1.4 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.2 | 2.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 1.0 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.2 | 3.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 0.7 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.2 | 1.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 14.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 1.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.2 | 2.7 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 1.8 | GO:0071546 | pi-body(GO:0071546) |
| 0.2 | 2.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 3.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 2.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 2.9 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 0.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 1.5 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 11.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.5 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 1.5 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 0.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.7 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.6 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.4 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 0.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.7 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.7 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 1.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 8.3 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 2.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.2 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.1 | 1.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 3.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 6.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 3.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 4.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.5 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 1.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 3.5 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.8 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.7 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 2.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 2.6 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.1 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 2.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 3.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 4.4 | GO:0031968 | organelle outer membrane(GO:0031968) |
| 0.0 | 0.6 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 5.2 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.8 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.1 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 6.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 6.3 | GO:0031012 | extracellular matrix(GO:0031012) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 1.2 | 3.5 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.9 | 4.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.7 | 3.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.7 | 2.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.7 | 2.8 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.7 | 6.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.7 | 6.6 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.6 | 1.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.5 | 6.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.5 | 1.5 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.5 | 2.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.5 | 1.5 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.5 | 2.3 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.4 | 1.7 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.4 | 1.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.4 | 1.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.4 | 2.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 1.5 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.3 | 1.4 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.3 | 4.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.3 | 2.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 2.6 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 1.8 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.2 | 4.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 2.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 3.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 1.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.2 | 3.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 2.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 0.9 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.2 | 2.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 2.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 0.5 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.2 | 0.5 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.2 | 1.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 9.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 3.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 3.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 1.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 1.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 9.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 1.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.4 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.1 | 1.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.8 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.4 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.1 | 2.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 3.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.7 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 0.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.3 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.1 | 0.8 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.3 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.1 | 10.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 2.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.6 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.7 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.3 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.1 | 0.6 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 3.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 12.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.7 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 3.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.7 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.3 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 3.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 2.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 1.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.3 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.1 | 1.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.6 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 2.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 1.6 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 1.7 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.2 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 1.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.2 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 0.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 5.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 3.0 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 2.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0050509 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 2.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 3.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.4 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 1.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.7 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 2.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 3.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 3.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 6.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 3.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.3 | GO:0097027 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 1.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 6.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.3 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 1.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 4.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 7.2 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 2.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.9 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.1 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 7.7 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.2 | 9.4 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.5 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.0 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.3 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.1 | 10.6 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.1 | 6.1 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.3 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 3.0 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.2 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.5 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.1 | 0.8 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.8 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.1 | 3.8 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 2.2 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.1 | 2.5 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.1 | 0.7 | PID_S1P_S1P2_PATHWAY | S1P2 pathway |
| 0.1 | 1.9 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 3.5 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.0 | 1.3 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.5 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.0 | 2.1 | PID_AMB2_NEUTROPHILS_PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.0 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 4.3 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | PID_IGF1_PATHWAY | IGF1 pathway |
| 0.0 | 0.6 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.5 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.1 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.5 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 1.7 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.6 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.2 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.0 | 0.5 | PID_NFKAPPAB_ATYPICAL_PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 2.1 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.0 | 0.8 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.4 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 7.1 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.7 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
| 0.0 | 0.4 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.7 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.5 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.9 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.0 | 0.3 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.2 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.1 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.9 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.6 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.3 | 9.8 | REACTOME_GRB2_SOS_PROVIDES_LINKAGE_TO_MAPK_SIGNALING_FOR_INTERGRINS_ | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.3 | 4.3 | REACTOME_REVERSIBLE_HYDRATION_OF_CARBON_DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 3.8 | REACTOME_TRYPTOPHAN_CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 1.5 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 2.2 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 4.5 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.1 | 9.0 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.9 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.5 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 4.0 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.5 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 2.3 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.5 | REACTOME_SPRY_REGULATION_OF_FGF_SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 3.5 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.1 | 2.0 | REACTOME_HYALURONAN_METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 2.9 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.2 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.3 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.7 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.2 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.4 | REACTOME_ER_PHAGOSOME_PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 3.4 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.1 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 3.3 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.6 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.9 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.5 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME_NFKB_IS_ACTIVATED_AND_SIGNALS_SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 1.6 | REACTOME_INTEGRIN_ALPHAIIB_BETA3_SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.4 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME_SIGNALING_BY_FGFR3_MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.5 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.1 | REACTOME_CA_DEPENDENT_EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 1.1 | REACTOME_PYRUVATE_METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 1.5 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 4.6 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME_GLUCAGON_SIGNALING_IN_METABOLIC_REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.6 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.6 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.7 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.5 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.7 | REACTOME_RIP_MEDIATED_NFKB_ACTIVATION_VIA_DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 2.4 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.1 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME_TRANS_GOLGI_NETWORK_VESICLE_BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.6 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 5.5 | REACTOME_METABOLISM_OF_AMINO_ACIDS_AND_DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.8 | REACTOME_SIGNALING_BY_PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.8 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.6 | REACTOME_CIRCADIAN_CLOCK | Genes involved in Circadian Clock |
| 0.0 | 1.1 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.3 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.0 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.6 | REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME_AUTODEGRADATION_OF_CDH1_BY_CDH1_APC_C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.4 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.7 | REACTOME_MUSCLE_CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.7 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.7 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.1 | REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.2 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME_INHIBITION_OF_INSULIN_SECRETION_BY_ADRENALINE_NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |