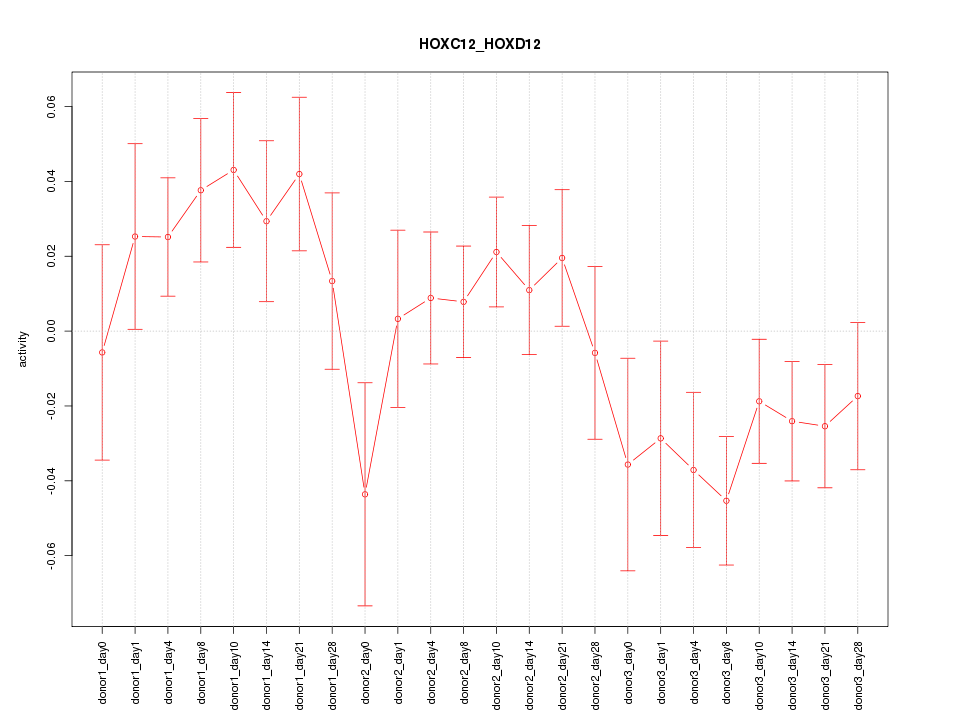
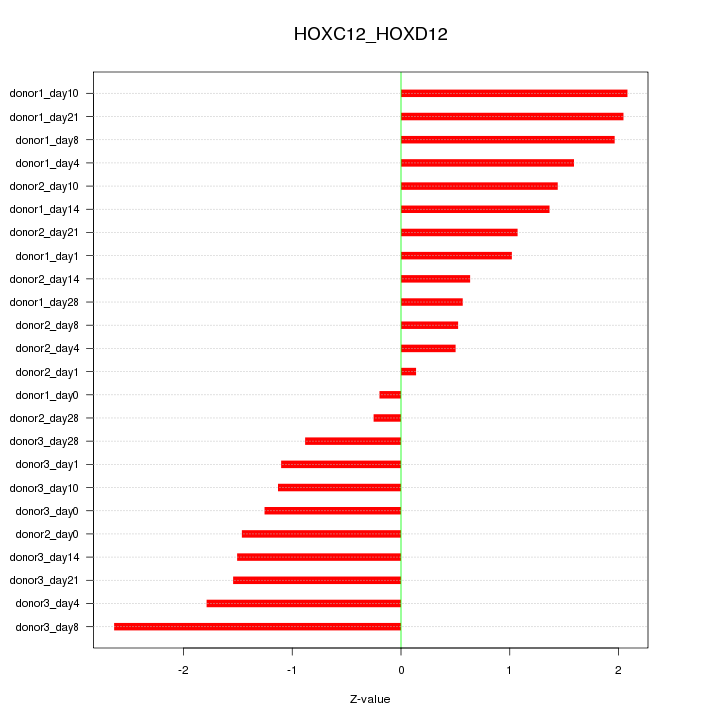
Motif ID: HOXC12_HOXD12
Z-value: 1.357
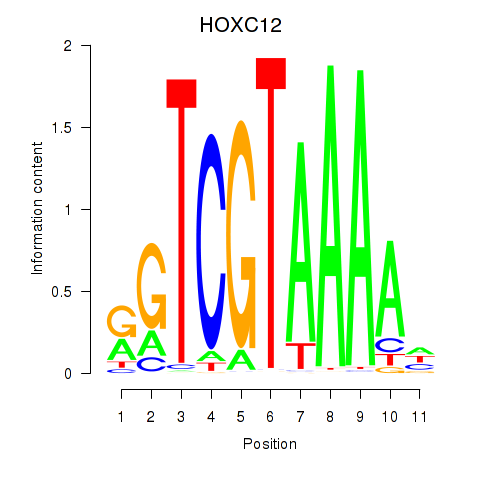
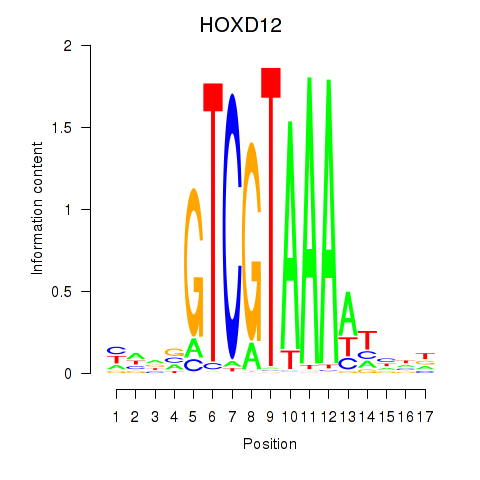
Transcription factors associated with HOXC12_HOXD12:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HOXC12 | ENSG00000123407.3 | HOXC12 |
| HOXD12 | ENSG00000170178.5 | HOXD12 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD12 | hg19_v2_chr2_+_176964458_176964540 | -0.56 | 4.2e-03 | Click! |
| HOXC12 | hg19_v2_chr12_+_54348618_54348693 | -0.08 | 7.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.4 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.7 | 2.1 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.6 | 2.3 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.4 | 1.1 | GO:0006174 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.4 | 1.1 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.4 | 1.8 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.3 | 4.5 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.3 | 10.4 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.3 | 0.8 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.2 | 0.7 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.2 | 3.2 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.2 | 0.7 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.2 | 1.0 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.2 | 0.9 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 1.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 1.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.6 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) seminal vesicle development(GO:0061107) |
| 0.1 | 0.6 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.9 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.9 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 20.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.4 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.1 | 0.8 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 2.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.6 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 1.8 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 3.3 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 0.6 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 0.4 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.7 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.1 | 0.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 1.5 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.4 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.1 | 2.3 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 1.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 1.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 2.5 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 2.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.4 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.5 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 1.2 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.4 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.7 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.5 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.5 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.8 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 1.3 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 1.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) maintenance of Golgi location(GO:0051684) |
| 0.0 | 1.3 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.7 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) |
| 0.0 | 0.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 1.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.8 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 0.1 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.9 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.8 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 21.4 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 0.9 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.2 | 0.9 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 3.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.2 | 3.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 0.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 1.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 2.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.5 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.0 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.3 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 0.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.5 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.8 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 0.3 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.4 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 2.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.2 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.3 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 1.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 2.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.3 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 1.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 1.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.5 | 3.2 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.3 | 2.9 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.3 | 1.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.3 | 1.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.3 | 2.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 20.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.2 | 0.7 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.2 | 1.8 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 2.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 0.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.2 | 0.9 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.2 | 0.5 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.1 | 1.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 1.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.9 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 1.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.1 | 0.5 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 2.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.4 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.4 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 1.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.9 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 4.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.4 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.4 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 1.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 1.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 1.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 1.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 2.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.8 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.0 | 1.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.3 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.9 | PID_TRAIL_PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.8 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 0.6 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.2 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.3 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.8 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID_FAS_PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.9 | PID_ENDOTHELIN_PATHWAY | Endothelins |
| 0.0 | 0.6 | PID_ATM_PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.3 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 20.4 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 3.3 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.4 | REACTOME_RECYCLING_OF_BILE_ACIDS_AND_SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.8 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.0 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.8 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.9 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.9 | REACTOME_EICOSANOID_LIGAND_BINDING_RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 4.5 | REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.8 | REACTOME_STEROID_HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.8 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.8 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.1 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.8 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.9 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.5 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.3 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 0.8 | REACTOME_AUTODEGRADATION_OF_CDH1_BY_CDH1_APC_C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.2 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 5.5 | REACTOME_GENERIC_TRANSCRIPTION_PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.3 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME_SIGNALING_BY_NOTCH4 | Genes involved in Signaling by NOTCH4 |