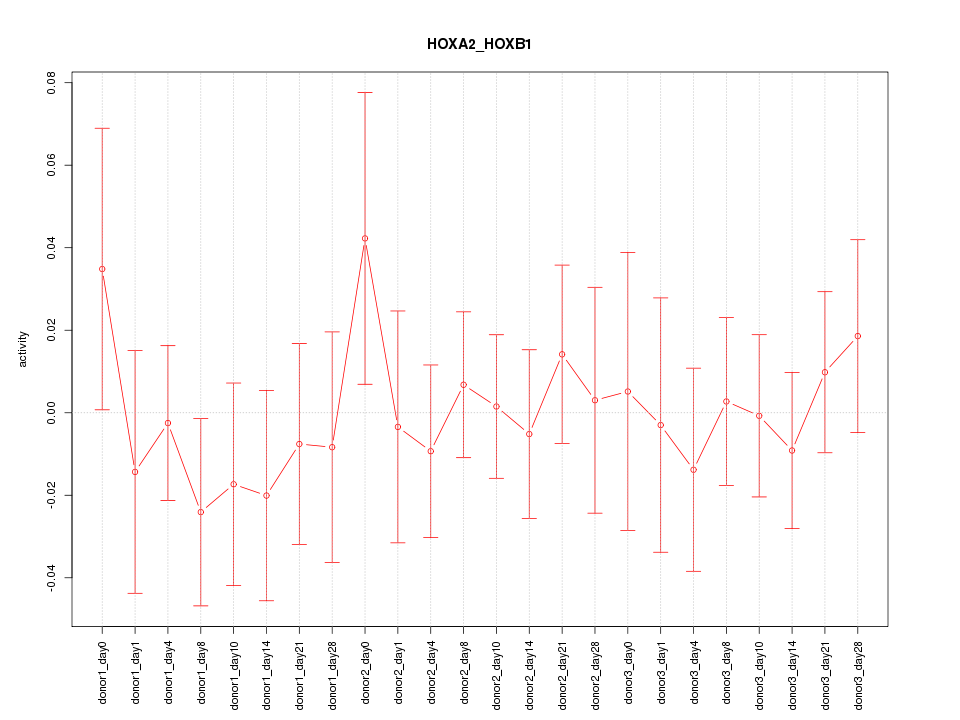
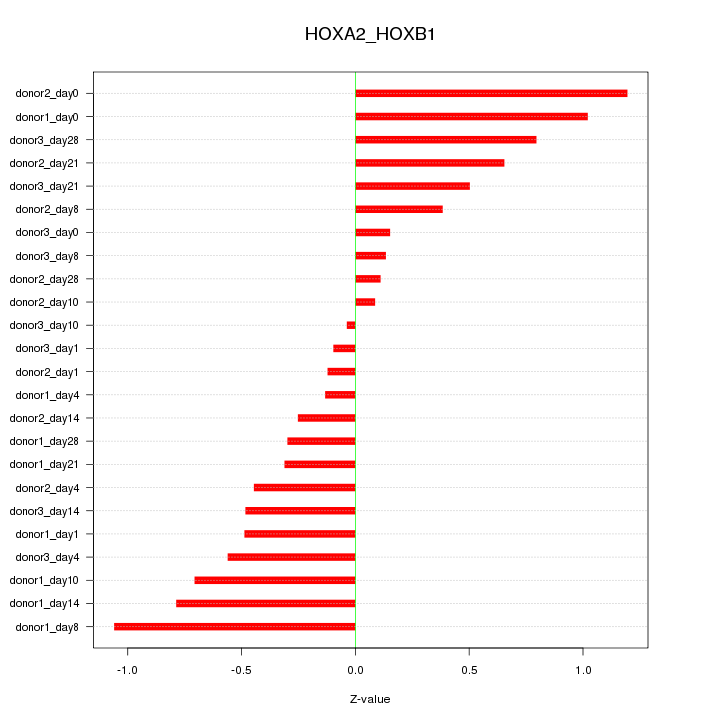
Motif ID: HOXA2_HOXB1
Z-value: 0.560
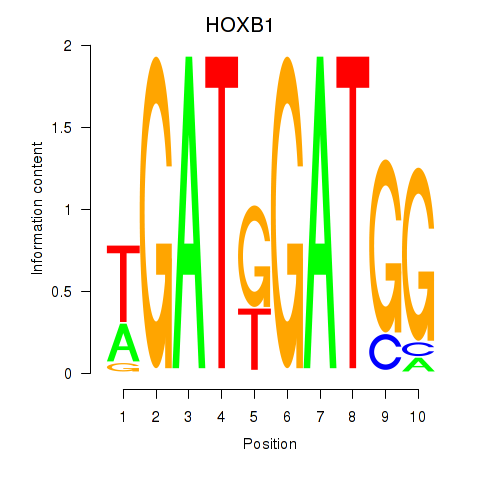
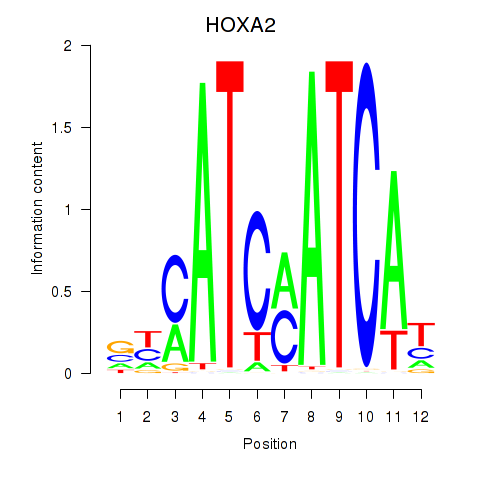
Transcription factors associated with HOXA2_HOXB1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HOXA2 | ENSG00000105996.5 | HOXA2 |
| HOXB1 | ENSG00000120094.6 | HOXB1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA2 | hg19_v2_chr7_-_27142290_27142430 | 0.20 | 3.4e-01 | Click! |
| HOXB1 | hg19_v2_chr17_-_46608272_46608385 | 0.02 | 9.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 | 1.6 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.4 | 1.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.3 | 4.8 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.3 | 1.4 | GO:1903285 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.3 | 1.3 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.2 | 0.7 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.2 | 1.2 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 1.3 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 1.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.3 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.3 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.4 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.0 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.2 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.2 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.2 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.7 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.3 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.3 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.0 | 0.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.6 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.0 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.1 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 1.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.7 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 4.8 | GO:0005938 | cell cortex(GO:0005938) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.3 | 4.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.3 | 1.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.3 | 1.0 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.2 | 1.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.3 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.1 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.3 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 1.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.3 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.5 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.6 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.4 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.2 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.0 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME_IRAK2_MEDIATED_ACTIVATION_OF_TAK1_COMPLEX_UPON_TLR7_8_OR_9_STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.6 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.4 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME_ENDOGENOUS_STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.6 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |