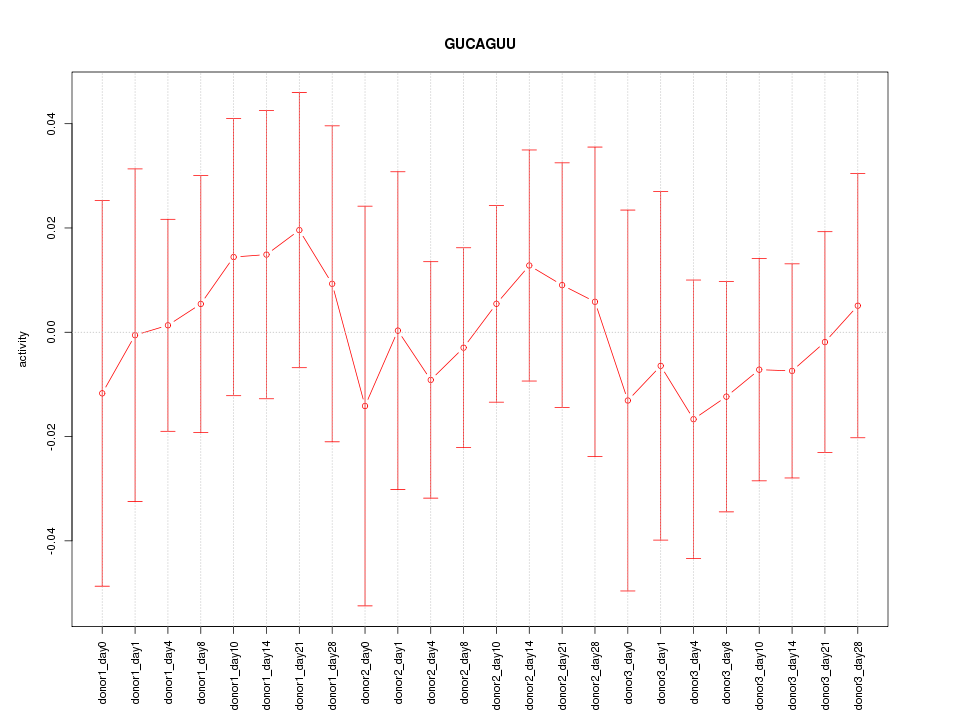
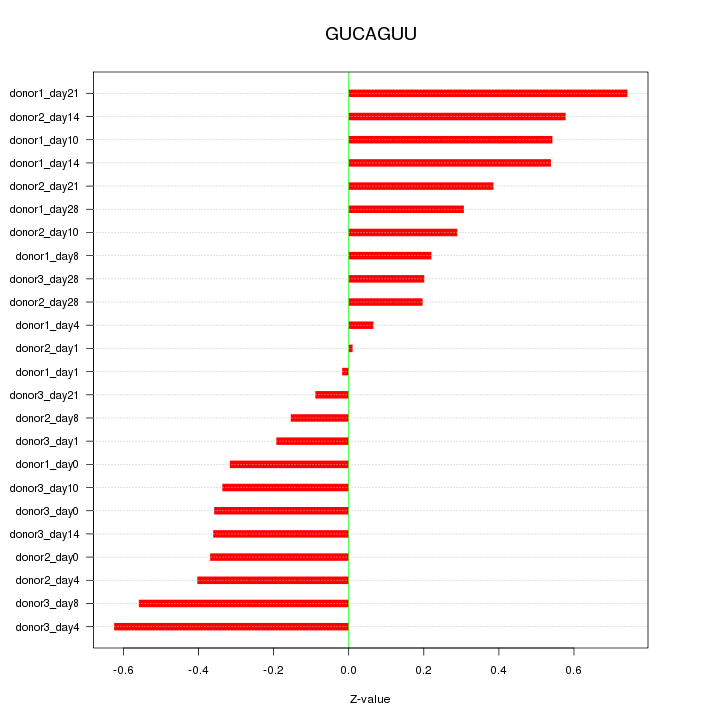
| 0.2 |
0.6 |
GO:0035283 |
rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 |
0.3 |
GO:0015993 |
L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 |
0.3 |
GO:1904158 |
axonemal central apparatus assembly(GO:1904158) |
| 0.1 |
0.2 |
GO:0071284 |
cellular response to lead ion(GO:0071284) |
| 0.0 |
0.2 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 |
0.1 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 |
0.1 |
GO:1902310 |
regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 |
0.3 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.0 |
0.1 |
GO:0035669 |
TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 |
0.1 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 0.0 |
0.1 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.0 |
0.1 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.0 |
0.1 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 |
0.1 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.0 |
0.2 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.1 |
GO:1990034 |
calcium ion export from cell(GO:1990034) |
| 0.0 |
0.1 |
GO:2000645 |
negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 |
0.1 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.0 |
GO:0072709 |
cellular response to sorbitol(GO:0072709) |
| 0.0 |
0.1 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.1 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.1 |
GO:0001692 |
histamine metabolic process(GO:0001692) |
| 0.0 |
0.1 |
GO:0070417 |
cellular response to cold(GO:0070417) |
| 0.0 |
0.1 |
GO:0007042 |
lysosomal lumen acidification(GO:0007042) |
| 0.0 |
0.1 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 |
0.1 |
GO:0043634 |
polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.0 |
0.3 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 |
0.0 |
GO:0006542 |
glutamine biosynthetic process(GO:0006542) |
| 0.0 |
0.1 |
GO:1902741 |
interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 |
0.0 |
GO:0015883 |
FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.0 |
0.1 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |