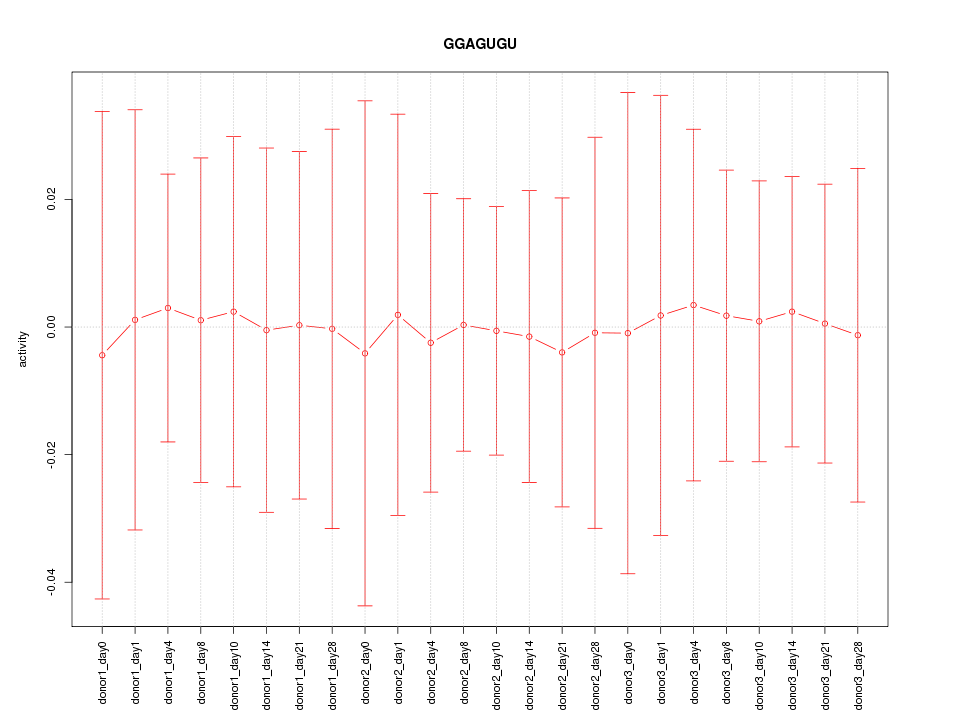
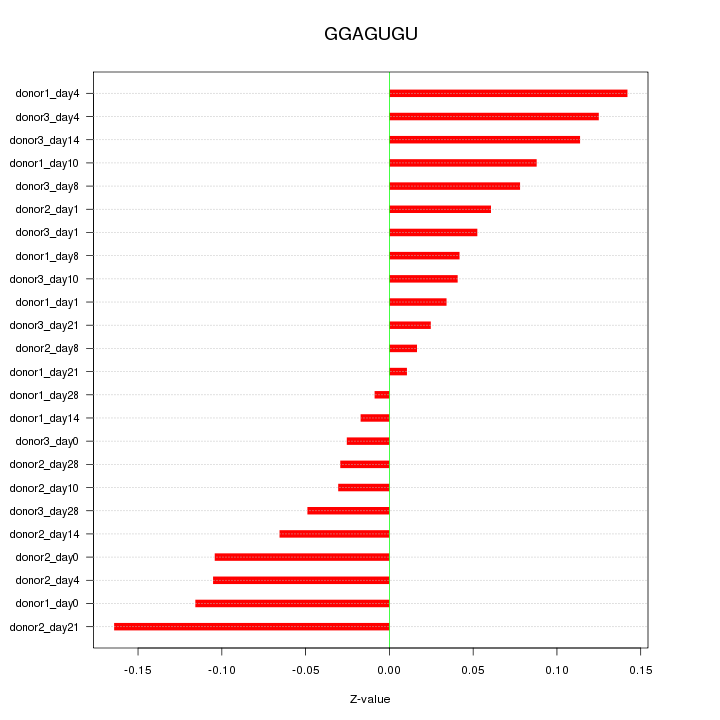
|
chr7_+_94285637
|
0.054
|
ENST00000482108.1
ENST00000488574.1
|
PEG10
|
paternally expressed 10
|
|
chr7_-_95225768
|
0.054
|
ENST00000005178.5
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4
|
|
chr8_+_102504651
|
0.046
|
ENST00000251808.3
ENST00000521085.1
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr8_-_29208183
|
0.037
|
ENST00000240100.2
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr19_+_46800289
|
0.025
|
ENST00000377670.4
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr19_-_2456922
|
0.023
|
ENST00000582871.1
ENST00000325327.3
|
LMNB2
|
lamin B2
|
|
chr1_+_28586006
|
0.023
|
ENST00000253063.3
|
SESN2
|
sestrin 2
|
|
chr1_-_245027833
|
0.022
|
ENST00000444376.2
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chr8_+_16884740
|
0.020
|
ENST00000318063.5
|
MICU3
|
mitochondrial calcium uptake family, member 3
|
|
chr20_-_31071239
|
0.019
|
ENST00000359676.5
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr3_+_9773409
|
0.018
|
ENST00000433861.2
ENST00000424362.1
ENST00000383829.2
ENST00000302054.3
ENST00000420291.1
|
BRPF1
|
bromodomain and PHD finger containing, 1
|
|
chr6_-_45983581
|
0.017
|
ENST00000339561.6
|
CLIC5
|
chloride intracellular channel 5
|
|
chr16_-_31076332
|
0.014
|
ENST00000539836.3
ENST00000535577.1
ENST00000442862.2
|
ZNF668
|
zinc finger protein 668
|
|
chr1_-_205782304
|
0.013
|
ENST00000367137.3
|
SLC41A1
|
solute carrier family 41 (magnesium transporter), member 1
|
|
chr16_+_69599861
|
0.012
|
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr7_+_100318423
|
0.010
|
ENST00000252723.2
|
EPO
|
erythropoietin
|
|
chr10_+_70480963
|
0.009
|
ENST00000265872.6
ENST00000535016.1
ENST00000538031.1
ENST00000543719.1
ENST00000539539.1
ENST00000543225.1
ENST00000536012.1
ENST00000494903.2
|
CCAR1
|
cell division cycle and apoptosis regulator 1
|
|
chr2_-_219925189
|
0.008
|
ENST00000295731.6
|
IHH
|
indian hedgehog
|
|
chr4_-_146859623
|
0.008
|
ENST00000379448.4
ENST00000513320.1
|
ZNF827
|
zinc finger protein 827
|
|
chr4_-_103266219
|
0.008
|
ENST00000394833.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chr10_-_23003460
|
0.007
|
ENST00000376573.4
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha
|
|
chr19_+_41816053
|
0.006
|
ENST00000269967.3
|
CCDC97
|
coiled-coil domain containing 97
|
|
chr6_+_72596604
|
0.006
|
ENST00000348717.5
ENST00000517960.1
ENST00000518273.1
ENST00000522291.1
ENST00000521978.1
ENST00000520567.1
ENST00000264839.7
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_+_16062820
|
0.005
|
ENST00000294454.5
|
SLC25A34
|
solute carrier family 25, member 34
|
|
chr3_+_47324424
|
0.005
|
ENST00000437353.1
ENST00000232766.5
ENST00000455924.2
|
KLHL18
|
kelch-like family member 18
|
|
chrX_-_135056216
|
0.005
|
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1
|
|
chr1_-_244615425
|
0.005
|
ENST00000366535.3
|
ADSS
|
adenylosuccinate synthase
|
|
chr3_-_187009646
|
0.005
|
ENST00000296280.6
ENST00000392470.2
ENST00000169293.6
ENST00000439271.1
ENST00000392472.2
ENST00000392475.2
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor)
|
|
chr6_+_152011628
|
0.005
|
ENST00000404742.1
ENST00000440973.1
|
ESR1
|
estrogen receptor 1
|
|
chr15_-_83316254
|
0.005
|
ENST00000567678.1
ENST00000450751.2
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr5_-_11904152
|
0.004
|
ENST00000304623.8
ENST00000458100.2
|
CTNND2
|
catenin (cadherin-associated protein), delta 2
|
|
chr3_+_183967409
|
0.004
|
ENST00000324557.4
ENST00000402825.3
|
ECE2
|
endothelin converting enzyme 2
|
|
chr14_-_77787198
|
0.004
|
ENST00000261534.4
|
POMT2
|
protein-O-mannosyltransferase 2
|
|
chr17_+_41857793
|
0.004
|
ENST00000449302.3
|
C17orf105
|
chromosome 17 open reading frame 105
|
|
chr5_+_179125907
|
0.004
|
ENST00000247461.4
ENST00000452673.2
ENST00000502498.1
ENST00000507307.1
ENST00000513246.1
ENST00000502673.1
ENST00000506654.1
ENST00000512607.2
ENST00000510810.1
|
CANX
|
calnexin
|
|
chr4_+_145567173
|
0.003
|
ENST00000296575.3
|
HHIP
|
hedgehog interacting protein
|
|
chr10_-_61666267
|
0.003
|
ENST00000263102.6
|
CCDC6
|
coiled-coil domain containing 6
|
|
chr21_-_40685477
|
0.003
|
ENST00000342449.3
|
BRWD1
|
bromodomain and WD repeat domain containing 1
|
|
chr4_-_76598296
|
0.003
|
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr5_-_141392538
|
0.002
|
ENST00000503794.1
ENST00000510194.1
ENST00000504424.1
ENST00000513454.1
ENST00000458112.2
ENST00000542860.1
ENST00000503229.1
ENST00000500692.2
ENST00000311337.6
ENST00000504139.1
ENST00000505689.1
|
GNPDA1
|
glucosamine-6-phosphate deaminase 1
|
|
chr10_-_32345305
|
0.002
|
ENST00000302418.4
|
KIF5B
|
kinesin family member 5B
|
|
chr12_-_56694142
|
0.002
|
ENST00000550655.1
ENST00000548567.1
ENST00000551430.2
ENST00000351328.3
|
CS
|
citrate synthase
|
|
chr5_+_125758813
|
0.002
|
ENST00000285689.3
ENST00000515200.1
|
GRAMD3
|
GRAM domain containing 3
|
|
chr7_+_114055052
|
0.002
|
ENST00000462331.1
ENST00000408937.3
ENST00000403559.4
ENST00000350908.4
ENST00000393498.2
ENST00000393495.3
ENST00000378237.3
ENST00000393489.3
|
FOXP2
|
forkhead box P2
|
|
chr20_+_32581452
|
0.002
|
ENST00000375114.3
ENST00000448364.1
|
RALY
|
RALY heterogeneous nuclear ribonucleoprotein
|
|
chr2_+_14772810
|
0.001
|
ENST00000295092.2
ENST00000331243.4
|
FAM84A
|
family with sequence similarity 84, member A
|
|
chr2_-_86948245
|
0.001
|
ENST00000439940.2
ENST00000604011.1
|
CHMP3
RNF103-CHMP3
|
charged multivesicular body protein 3
RNF103-CHMP3 readthrough
|
|
chr6_-_91006461
|
0.001
|
ENST00000257749.4
ENST00000343122.3
ENST00000406998.2
ENST00000453877.1
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr1_-_72748417
|
0.001
|
ENST00000357731.5
|
NEGR1
|
neuronal growth regulator 1
|
|
chr1_+_25071848
|
0.001
|
ENST00000374379.4
|
CLIC4
|
chloride intracellular channel 4
|
|
chr8_+_145159376
|
0.001
|
ENST00000322428.5
|
MAF1
|
MAF1 homolog (S. cerevisiae)
|
|
chr16_-_30381580
|
0.001
|
ENST00000409939.3
|
TBC1D10B
|
TBC1 domain family, member 10B
|
|
chr7_-_33102338
|
0.001
|
ENST00000610140.1
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA
|
|
chr19_-_6279932
|
0.000
|
ENST00000252674.7
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1
|
|
chr19_-_47291843
|
0.000
|
ENST00000542575.2
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5
|