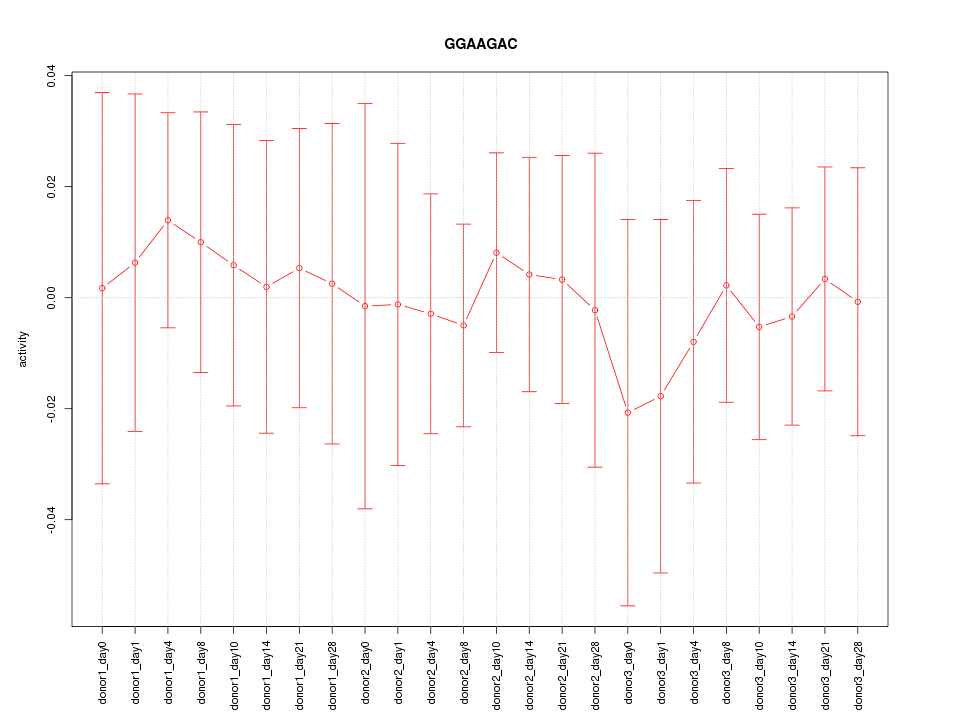
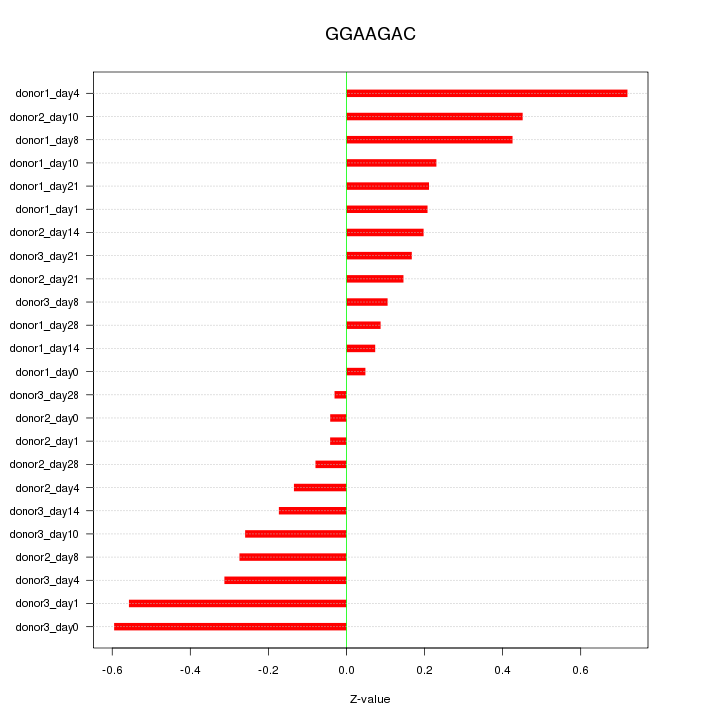
|
chr11_-_115375107
|
0.412
|
ENST00000545380.1
ENST00000452722.3
ENST00000537058.1
ENST00000536727.1
ENST00000542447.2
ENST00000331581.6
|
CADM1
|
cell adhesion molecule 1
|
|
chr14_-_90085458
|
0.365
|
ENST00000345097.4
ENST00000555855.1
ENST00000555353.1
|
FOXN3
|
forkhead box N3
|
|
chr1_-_217262969
|
0.299
|
ENST00000361525.3
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr3_-_18466787
|
0.268
|
ENST00000338745.6
ENST00000450898.1
|
SATB1
|
SATB homeobox 1
|
|
chr13_-_110438914
|
0.263
|
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2
|
|
chr17_+_68165657
|
0.245
|
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr5_-_111093406
|
0.227
|
ENST00000379671.3
|
NREP
|
neuronal regeneration related protein
|
|
chr13_+_115079949
|
0.226
|
ENST00000361283.1
|
CHAMP1
|
chromosome alignment maintaining phosphoprotein 1
|
|
chr18_-_53255766
|
0.219
|
ENST00000566286.1
ENST00000564999.1
ENST00000566279.1
ENST00000354452.3
ENST00000356073.4
|
TCF4
|
transcription factor 4
|
|
chr1_+_61547894
|
0.216
|
ENST00000403491.3
|
NFIA
|
nuclear factor I/A
|
|
chr19_+_45504688
|
0.213
|
ENST00000221452.8
ENST00000540120.1
ENST00000505236.1
|
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B
|
|
chr1_+_162351503
|
0.211
|
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226
|
|
chr12_-_56727487
|
0.202
|
ENST00000548043.1
ENST00000425394.2
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr20_-_33460621
|
0.197
|
ENST00000427420.1
ENST00000336431.5
|
GGT7
|
gamma-glutamyltransferase 7
|
|
chr17_+_29718642
|
0.186
|
ENST00000325874.8
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr19_+_10527449
|
0.182
|
ENST00000592685.1
ENST00000380702.2
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr7_-_75368248
|
0.175
|
ENST00000434438.2
ENST00000336926.6
|
HIP1
|
huntingtin interacting protein 1
|
|
chr4_+_72204755
|
0.175
|
ENST00000512686.1
ENST00000340595.3
|
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4
|
|
chr3_+_29322803
|
0.169
|
ENST00000396583.3
ENST00000383767.2
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr11_-_73309228
|
0.168
|
ENST00000356467.4
ENST00000064778.4
|
FAM168A
|
family with sequence similarity 168, member A
|
|
chr20_+_5986727
|
0.168
|
ENST00000378863.4
|
CRLS1
|
cardiolipin synthase 1
|
|
chr15_+_44719394
|
0.167
|
ENST00000260327.4
ENST00000396780.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2
|
|
chr3_+_45636219
|
0.166
|
ENST00000273317.4
|
LIMD1
|
LIM domains containing 1
|
|
chr7_+_77325738
|
0.165
|
ENST00000334955.8
|
RSBN1L
|
round spermatid basic protein 1-like
|
|
chr14_-_70883708
|
0.165
|
ENST00000256366.4
|
SYNJ2BP
|
synaptojanin 2 binding protein
|
|
chr5_+_137688285
|
0.165
|
ENST00000314358.5
|
KDM3B
|
lysine (K)-specific demethylase 3B
|
|
chr8_+_107670064
|
0.160
|
ENST00000312046.6
|
OXR1
|
oxidation resistance 1
|
|
chrX_+_108780062
|
0.157
|
ENST00000372106.1
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr4_+_184826418
|
0.153
|
ENST00000308497.4
ENST00000438269.1
|
STOX2
|
storkhead box 2
|
|
chr4_+_129730779
|
0.152
|
ENST00000226319.6
|
PHF17
|
jade family PHD finger 1
|
|
chr1_-_46598284
|
0.148
|
ENST00000423209.1
ENST00000262741.5
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr19_-_47616992
|
0.143
|
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chr1_+_229440129
|
0.141
|
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related)
|
|
chr2_-_97535708
|
0.141
|
ENST00000305476.5
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C
|
|
chr1_+_118148556
|
0.138
|
ENST00000369448.3
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr1_+_110082487
|
0.138
|
ENST00000527748.1
|
GPR61
|
G protein-coupled receptor 61
|
|
chr1_+_89990431
|
0.132
|
ENST00000330947.2
ENST00000358200.4
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr5_-_59189545
|
0.131
|
ENST00000340635.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_-_12705600
|
0.130
|
ENST00000542177.1
ENST00000442415.2
ENST00000251849.4
|
RAF1
|
v-raf-1 murine leukemia viral oncogene homolog 1
|
|
chr8_+_26435359
|
0.130
|
ENST00000311151.5
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr20_-_3996036
|
0.121
|
ENST00000336095.6
|
RNF24
|
ring finger protein 24
|
|
chr15_+_57210818
|
0.118
|
ENST00000438423.2
ENST00000267811.5
ENST00000452095.2
ENST00000559609.1
ENST00000333725.5
|
TCF12
|
transcription factor 12
|
|
chr7_-_10979750
|
0.118
|
ENST00000339600.5
|
NDUFA4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa
|
|
chr1_+_184356188
|
0.112
|
ENST00000235307.6
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr10_+_92980517
|
0.110
|
ENST00000336126.5
|
PCGF5
|
polycomb group ring finger 5
|
|
chr6_-_16761678
|
0.109
|
ENST00000244769.4
ENST00000436367.1
|
ATXN1
|
ataxin 1
|
|
chr2_-_213403565
|
0.108
|
ENST00000342788.4
ENST00000436443.1
|
ERBB4
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4
|
|
chr9_-_123476719
|
0.107
|
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr7_+_138818490
|
0.105
|
ENST00000430935.1
ENST00000495038.1
ENST00000474035.2
ENST00000478836.2
ENST00000464848.1
ENST00000343187.4
|
TTC26
|
tetratricopeptide repeat domain 26
|
|
chr9_+_104296122
|
0.104
|
ENST00000389120.3
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase
|
|
chr7_+_139026057
|
0.103
|
ENST00000541515.3
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae)
|
|
chr3_-_88108192
|
0.103
|
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1
|
|
chr18_+_9136758
|
0.102
|
ENST00000383440.2
ENST00000262126.4
ENST00000577992.1
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr11_+_45868957
|
0.095
|
ENST00000443527.2
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr19_-_2721412
|
0.091
|
ENST00000323469.4
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr17_+_5390220
|
0.086
|
ENST00000381165.3
|
MIS12
|
MIS12 kinetochore complex component
|
|
chr17_-_16118835
|
0.086
|
ENST00000582357.1
ENST00000436828.1
ENST00000411510.1
ENST00000268712.3
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr21_+_35445827
|
0.085
|
ENST00000381151.3
ENST00000608209.1
|
SLC5A3
SLC5A3
|
solute carrier family 5 (sodium/myo-inositol cotransporter), member 3
sodium/myo-inositol cotransporter
|
|
chr7_-_35734730
|
0.080
|
ENST00000396081.1
ENST00000311350.3
|
HERPUD2
|
HERPUD family member 2
|
|
chr17_-_56406117
|
0.078
|
ENST00000268893.6
ENST00000355701.3
|
BZRAP1
|
benzodiazepine receptor (peripheral) associated protein 1
|
|
chr1_-_114355083
|
0.078
|
ENST00000261441.5
|
RSBN1
|
round spermatid basic protein 1
|
|
chr2_-_161350305
|
0.078
|
ENST00000348849.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr18_-_45663666
|
0.077
|
ENST00000535628.2
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr1_+_206557366
|
0.075
|
ENST00000414007.1
ENST00000419187.2
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2
|
|
chr1_+_27561007
|
0.074
|
ENST00000319394.3
|
WDTC1
|
WD and tetratricopeptide repeats 1
|
|
chr12_+_60083118
|
0.073
|
ENST00000261187.4
ENST00000543448.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7
|
|
chr17_+_25799008
|
0.072
|
ENST00000583370.1
ENST00000398988.3
ENST00000268763.6
|
KSR1
|
kinase suppressor of ras 1
|
|
chr10_-_94333784
|
0.069
|
ENST00000265986.6
|
IDE
|
insulin-degrading enzyme
|
|
chr4_-_99851766
|
0.067
|
ENST00000450253.2
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr1_+_231297798
|
0.066
|
ENST00000444294.3
|
TRIM67
|
tripartite motif containing 67
|
|
chr3_-_52931557
|
0.066
|
ENST00000355083.5
ENST00000504329.1
|
TMEM110
TMEM110-MUSTN1
|
transmembrane protein 110
TMEM110-MUSTN1 readthrough
|
|
chr2_-_152955537
|
0.065
|
ENST00000201943.5
ENST00000539935.1
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr11_-_65816591
|
0.065
|
ENST00000312006.4
|
GAL3ST3
|
galactose-3-O-sulfotransferase 3
|
|
chr12_-_58240470
|
0.063
|
ENST00000548823.1
ENST00000398073.2
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2
|
|
chr11_-_46142948
|
0.063
|
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A
|
|
chr12_-_46766577
|
0.063
|
ENST00000256689.5
|
SLC38A2
|
solute carrier family 38, member 2
|
|
chr5_+_158690089
|
0.062
|
ENST00000296786.6
|
UBLCP1
|
ubiquitin-like domain containing CTD phosphatase 1
|
|
chr12_+_122150646
|
0.060
|
ENST00000449592.2
|
TMEM120B
|
transmembrane protein 120B
|
|
chr3_-_125094093
|
0.058
|
ENST00000484491.1
ENST00000492394.1
ENST00000471196.1
ENST00000468369.1
ENST00000544464.1
ENST00000485866.1
ENST00000360647.4
|
ZNF148
|
zinc finger protein 148
|
|
chr12_+_1929421
|
0.056
|
ENST00000543818.1
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2
|
|
chr10_+_72164135
|
0.055
|
ENST00000373218.4
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr20_+_30946106
|
0.054
|
ENST00000375687.4
ENST00000542461.1
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chrX_+_70752917
|
0.053
|
ENST00000373719.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase
|
|
chr8_-_57123815
|
0.052
|
ENST00000316981.3
ENST00000423799.2
ENST00000429357.2
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr3_-_112360116
|
0.052
|
ENST00000206423.3
ENST00000439685.2
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_+_28995231
|
0.052
|
ENST00000373816.1
|
GMEB1
|
glucocorticoid modulatory element binding protein 1
|
|
chr2_-_242212227
|
0.050
|
ENST00000427007.1
ENST00000458564.1
ENST00000452065.1
ENST00000427183.2
ENST00000426343.1
ENST00000422080.1
ENST00000449504.1
ENST00000449864.1
ENST00000391975.1
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr1_-_95392635
|
0.048
|
ENST00000538964.1
ENST00000394202.4
ENST00000370206.4
|
CNN3
|
calponin 3, acidic
|
|
chr10_-_112064665
|
0.047
|
ENST00000369603.5
|
SMNDC1
|
survival motor neuron domain containing 1
|
|
chr12_+_56661033
|
0.045
|
ENST00000433805.2
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr3_-_133969437
|
0.044
|
ENST00000460933.1
ENST00000296084.4
|
RYK
|
receptor-like tyrosine kinase
|
|
chr20_-_50159198
|
0.043
|
ENST00000371564.3
ENST00000396009.3
ENST00000610033.1
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr16_+_15068955
|
0.043
|
ENST00000396410.4
ENST00000569715.1
ENST00000450288.2
|
PDXDC1
|
pyridoxal-dependent decarboxylase domain containing 1
|
|
chr3_-_15901278
|
0.041
|
ENST00000399451.2
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr7_-_44924939
|
0.041
|
ENST00000395699.2
|
PURB
|
purine-rich element binding protein B
|
|
chr6_-_134639180
|
0.040
|
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_-_114790179
|
0.038
|
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chrX_+_150565653
|
0.038
|
ENST00000330374.6
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae)
|
|
chr12_-_14133053
|
0.038
|
ENST00000609686.1
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr11_+_85956182
|
0.038
|
ENST00000327320.4
ENST00000351625.6
ENST00000534595.1
|
EED
|
embryonic ectoderm development
|
|
chr7_-_139876812
|
0.037
|
ENST00000397560.2
|
JHDM1D
|
lysine (K)-specific demethylase 7A
|
|
chr7_+_75831181
|
0.034
|
ENST00000388802.4
ENST00000326382.8
|
SRRM3
|
serine/arginine repetitive matrix 3
|
|
chr6_-_3457256
|
0.033
|
ENST00000436008.2
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr19_+_3359561
|
0.032
|
ENST00000589123.1
ENST00000346156.5
ENST00000395111.3
ENST00000586919.1
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr10_-_46167722
|
0.031
|
ENST00000374366.3
ENST00000344646.5
|
ZFAND4
|
zinc finger, AN1-type domain 4
|
|
chr10_+_121652204
|
0.030
|
ENST00000369075.3
ENST00000543134.1
|
SEC23IP
|
SEC23 interacting protein
|
|
chr3_+_142720366
|
0.030
|
ENST00000493782.1
ENST00000397933.2
ENST00000473835.2
ENST00000493598.2
|
U2SURP
|
U2 snRNP-associated SURP domain containing
|
|
chr12_+_53774423
|
0.030
|
ENST00000426431.2
|
SP1
|
Sp1 transcription factor
|
|
chr13_+_27131887
|
0.029
|
ENST00000335327.5
|
WASF3
|
WAS protein family, member 3
|
|
chr12_+_65563329
|
0.028
|
ENST00000308330.2
|
LEMD3
|
LEM domain containing 3
|
|
chr16_-_18937726
|
0.028
|
ENST00000389467.3
ENST00000446231.2
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase
|
|
chr3_-_113415441
|
0.027
|
ENST00000491165.1
ENST00000316407.4
|
KIAA2018
|
KIAA2018
|
|
chr2_+_75185619
|
0.026
|
ENST00000483063.1
|
POLE4
|
polymerase (DNA-directed), epsilon 4, accessory subunit
|
|
chr19_+_19431490
|
0.025
|
ENST00000392313.6
ENST00000262815.8
ENST00000609122.1
|
MAU2
|
MAU2 sister chromatid cohesion factor
|
|
chr1_+_36348790
|
0.025
|
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1
|
|
chr1_+_206643787
|
0.024
|
ENST00000367120.3
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon
|
|
chr3_+_112280857
|
0.024
|
ENST00000492406.1
ENST00000468642.1
|
SLC35A5
|
solute carrier family 35, member A5
|
|
chr10_-_70287231
|
0.024
|
ENST00000609923.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16
|
|
chr17_-_42908155
|
0.023
|
ENST00000426548.1
ENST00000590758.1
ENST00000591424.1
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr7_-_42276612
|
0.023
|
ENST00000395925.3
ENST00000437480.1
|
GLI3
|
GLI family zinc finger 3
|
|
chr16_+_67143880
|
0.023
|
ENST00000219139.3
ENST00000566026.1
|
C16orf70
|
chromosome 16 open reading frame 70
|
|
chr1_+_162467595
|
0.023
|
ENST00000538489.1
ENST00000489294.1
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr5_+_61874562
|
0.022
|
ENST00000409534.1
ENST00000334994.5
|
IPO11
LRRC70
|
importin 11
leucine rich repeat containing 70
|
|
chr14_-_31495569
|
0.022
|
ENST00000357479.5
ENST00000355683.5
|
STRN3
|
striatin, calmodulin binding protein 3
|
|
chr14_-_92506371
|
0.021
|
ENST00000267622.4
|
TRIP11
|
thyroid hormone receptor interactor 11
|
|
chr12_-_95467356
|
0.021
|
ENST00000393101.3
ENST00000333003.5
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1
|
|
chr11_-_18656028
|
0.020
|
ENST00000336349.5
|
SPTY2D1
|
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae)
|
|
chr16_+_69599861
|
0.020
|
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr1_+_229406847
|
0.020
|
ENST00000366690.4
|
RAB4A
|
RAB4A, member RAS oncogene family
|
|
chr10_-_98945677
|
0.019
|
ENST00000266058.4
ENST00000371041.3
|
SLIT1
|
slit homolog 1 (Drosophila)
|
|
chr4_+_71768043
|
0.019
|
ENST00000502869.1
ENST00000309395.2
ENST00000396051.2
|
MOB1B
|
MOB kinase activator 1B
|
|
chr1_-_115053781
|
0.018
|
ENST00000358465.2
ENST00000369543.2
|
TRIM33
|
tripartite motif containing 33
|
|
chr17_+_40985407
|
0.018
|
ENST00000586114.1
ENST00000590720.1
ENST00000585805.1
ENST00000541124.1
ENST00000441946.2
ENST00000591152.1
ENST00000589469.1
ENST00000293362.3
ENST00000592169.1
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr13_+_48877895
|
0.018
|
ENST00000267163.4
|
RB1
|
retinoblastoma 1
|
|
chr5_+_17217669
|
0.017
|
ENST00000322611.3
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr2_+_178257372
|
0.017
|
ENST00000264167.4
ENST00000409888.1
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr10_-_88281494
|
0.017
|
ENST00000298767.5
|
WAPAL
|
wings apart-like homolog (Drosophila)
|
|
chr1_-_44497024
|
0.016
|
ENST00000372306.3
ENST00000372310.3
ENST00000475075.2
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr5_+_31532373
|
0.016
|
ENST00000325366.9
ENST00000355907.3
ENST00000507818.2
|
C5orf22
|
chromosome 5 open reading frame 22
|
|
chr11_-_70507901
|
0.016
|
ENST00000449833.2
ENST00000357171.3
ENST00000449116.2
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr15_+_77712993
|
0.015
|
ENST00000336216.4
ENST00000381714.3
ENST00000558651.1
|
HMG20A
|
high mobility group 20A
|
|
chr3_-_195270162
|
0.015
|
ENST00000438848.1
ENST00000328432.3
|
PPP1R2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2
|
|
chr1_+_43855560
|
0.015
|
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse)
|
|
chr21_-_40685477
|
0.015
|
ENST00000342449.3
|
BRWD1
|
bromodomain and WD repeat domain containing 1
|
|
chr16_+_3355472
|
0.015
|
ENST00000574298.1
|
ZNF75A
|
zinc finger protein 75a
|
|
chr1_-_207224307
|
0.015
|
ENST00000315927.4
|
YOD1
|
YOD1 deubiquitinase
|
|
chr20_+_3451650
|
0.014
|
ENST00000262919.5
|
ATRN
|
attractin
|
|
chr17_+_28804380
|
0.014
|
ENST00000225724.5
ENST00000451249.2
ENST00000467337.2
ENST00000581721.1
ENST00000414833.2
|
GOSR1
|
golgi SNAP receptor complex member 1
|
|
chr3_-_125313934
|
0.014
|
ENST00000296220.5
|
OSBPL11
|
oxysterol binding protein-like 11
|
|
chr9_-_14314066
|
0.013
|
ENST00000397575.3
|
NFIB
|
nuclear factor I/B
|
|
chr14_+_73603126
|
0.013
|
ENST00000557356.1
ENST00000556864.1
ENST00000556533.1
ENST00000556951.1
ENST00000557293.1
ENST00000553719.1
ENST00000553599.1
ENST00000556011.1
ENST00000394157.3
ENST00000357710.4
ENST00000324501.5
ENST00000560005.2
ENST00000555254.1
ENST00000261970.3
ENST00000344094.3
ENST00000554131.1
ENST00000557037.1
|
PSEN1
|
presenilin 1
|
|
chr6_-_107436473
|
0.013
|
ENST00000369042.1
|
BEND3
|
BEN domain containing 3
|
|
chr16_-_12009735
|
0.013
|
ENST00000439887.2
ENST00000434724.2
|
GSPT1
|
G1 to S phase transition 1
|
|
chrX_+_40944871
|
0.013
|
ENST00000378308.2
ENST00000324545.8
|
USP9X
|
ubiquitin specific peptidase 9, X-linked
|
|
chr4_-_114682936
|
0.012
|
ENST00000454265.2
ENST00000429180.1
ENST00000418639.2
ENST00000394526.2
ENST00000296402.5
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta
|
|
chr19_-_36523709
|
0.012
|
ENST00000592017.1
ENST00000360535.4
|
CLIP3
|
CAP-GLY domain containing linker protein 3
|
|
chr1_-_153895377
|
0.012
|
ENST00000368655.4
|
GATAD2B
|
GATA zinc finger domain containing 2B
|
|
chr3_+_150804676
|
0.012
|
ENST00000474524.1
ENST00000273432.4
|
MED12L
|
mediator complex subunit 12-like
|
|
chr12_-_49449107
|
0.011
|
ENST00000301067.7
|
KMT2D
|
lysine (K)-specific methyltransferase 2D
|
|
chr5_+_56469775
|
0.011
|
ENST00000424459.3
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr1_-_35325400
|
0.010
|
ENST00000521580.2
|
SMIM12
|
small integral membrane protein 12
|
|
chr13_-_72441315
|
0.010
|
ENST00000305425.4
ENST00000313174.7
ENST00000354591.4
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr7_+_106809406
|
0.010
|
ENST00000468410.1
ENST00000478930.1
ENST00000464009.1
ENST00000222574.4
|
HBP1
|
HMG-box transcription factor 1
|
|
chr4_-_103748880
|
0.010
|
ENST00000453744.2
ENST00000349311.8
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3
|
|
chr8_+_87354945
|
0.010
|
ENST00000517970.1
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1
|
|
chr2_-_201936302
|
0.010
|
ENST00000453765.1
ENST00000452799.1
ENST00000446678.1
ENST00000418596.3
|
FAM126B
|
family with sequence similarity 126, member B
|
|
chrX_+_107334895
|
0.010
|
ENST00000372232.3
ENST00000345734.3
ENST00000372254.3
|
ATG4A
|
autophagy related 4A, cysteine peptidase
|
|
chr16_+_16043406
|
0.009
|
ENST00000399410.3
ENST00000399408.2
ENST00000346370.5
ENST00000351154.5
ENST00000345148.5
ENST00000349029.5
|
ABCC1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 1
|
|
chr8_-_145559943
|
0.009
|
ENST00000332135.4
|
SCRT1
|
scratch family zinc finger 1
|
|
chr12_+_122064398
|
0.009
|
ENST00000330079.7
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1
|
|
chr2_+_234263120
|
0.009
|
ENST00000264057.2
ENST00000427930.1
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr10_-_115904361
|
0.009
|
ENST00000428953.1
ENST00000543782.1
|
C10orf118
|
chromosome 10 open reading frame 118
|
|
chr2_+_135676381
|
0.009
|
ENST00000537343.1
ENST00000295238.6
ENST00000264157.5
|
CCNT2
|
cyclin T2
|
|
chr2_+_139259324
|
0.008
|
ENST00000280098.4
|
SPOPL
|
speckle-type POZ protein-like
|
|
chr11_-_62521614
|
0.008
|
ENST00000527994.1
ENST00000394807.3
|
ZBTB3
|
zinc finger and BTB domain containing 3
|
|
chr1_+_193091080
|
0.008
|
ENST00000367435.3
|
CDC73
|
cell division cycle 73
|
|
chr12_+_93965451
|
0.007
|
ENST00000548537.1
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr10_+_81107216
|
0.006
|
ENST00000394579.3
ENST00000225174.3
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr4_+_15004165
|
0.006
|
ENST00000538197.1
ENST00000541112.1
ENST00000442003.2
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr11_-_75062730
|
0.006
|
ENST00000420843.2
ENST00000360025.3
|
ARRB1
|
arrestin, beta 1
|
|
chr1_-_160068465
|
0.006
|
ENST00000314485.7
ENST00000368086.1
|
IGSF8
|
immunoglobulin superfamily, member 8
|
|
chr16_+_85646763
|
0.006
|
ENST00000411612.1
ENST00000253458.7
|
GSE1
|
Gse1 coiled-coil protein
|
|
chr5_+_154238096
|
0.006
|
ENST00000517568.1
ENST00000524105.1
ENST00000285896.6
|
CNOT8
|
CCR4-NOT transcription complex, subunit 8
|
|
chr17_-_19771216
|
0.005
|
ENST00000395544.4
|
ULK2
|
unc-51 like autophagy activating kinase 2
|
|
chr7_+_139044621
|
0.005
|
ENST00000354926.4
|
C7orf55-LUC7L2
|
C7orf55-LUC7L2 readthrough
|
|
chr10_+_80828774
|
0.005
|
ENST00000334512.5
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr12_+_67663056
|
0.005
|
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1
|
|
chr6_+_111195973
|
0.005
|
ENST00000368885.3
ENST00000368882.3
ENST00000451850.2
ENST00000368877.5
|
AMD1
|
adenosylmethionine decarboxylase 1
|
|
chr15_-_65809581
|
0.004
|
ENST00000341861.5
|
DPP8
|
dipeptidyl-peptidase 8
|
|
chr3_+_180630090
|
0.004
|
ENST00000357559.4
ENST00000305586.7
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr3_-_33686743
|
0.004
|
ENST00000333778.6
ENST00000539981.1
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr20_+_49348081
|
0.004
|
ENST00000371610.2
|
PARD6B
|
par-6 family cell polarity regulator beta
|
|
chr7_-_71801980
|
0.004
|
ENST00000329008.5
|
CALN1
|
calneuron 1
|
|
chr3_-_52312636
|
0.004
|
ENST00000296490.3
|
WDR82
|
WD repeat domain 82
|
|
chr17_-_1465924
|
0.004
|
ENST00000573231.1
ENST00000576722.1
ENST00000576761.1
ENST00000576010.2
ENST00000313486.7
ENST00000539476.1
|
PITPNA
|
phosphatidylinositol transfer protein, alpha
|
|
chr12_+_56367697
|
0.004
|
ENST00000553116.1
ENST00000360299.5
ENST00000548068.1
ENST00000549915.1
ENST00000551459.1
ENST00000448789.2
|
RAB5B
|
RAB5B, member RAS oncogene family
|
|
chr14_-_45431091
|
0.003
|
ENST00000579157.1
ENST00000396128.4
ENST00000556500.1
|
KLHL28
|
kelch-like family member 28
|
|
chr7_+_148936732
|
0.003
|
ENST00000335870.2
|
ZNF212
|
zinc finger protein 212
|
|
chr8_+_42249418
|
0.003
|
ENST00000521158.1
ENST00000022615.4
|
VDAC3
|
voltage-dependent anion channel 3
|
|
chr4_-_87281224
|
0.003
|
ENST00000395169.3
ENST00000395161.2
|
MAPK10
|
mitogen-activated protein kinase 10
|