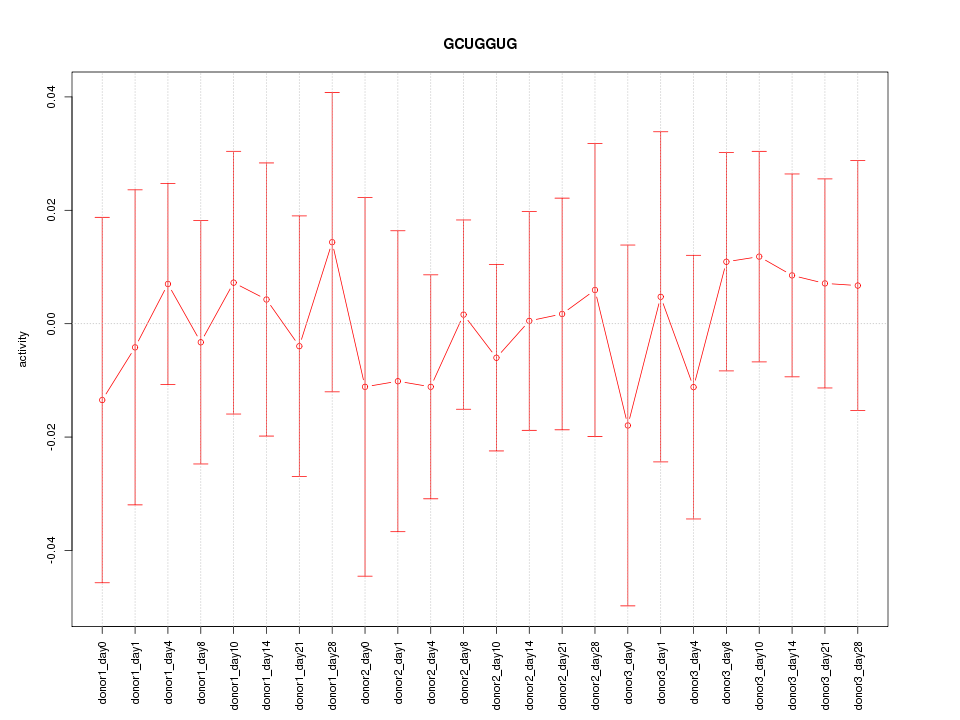
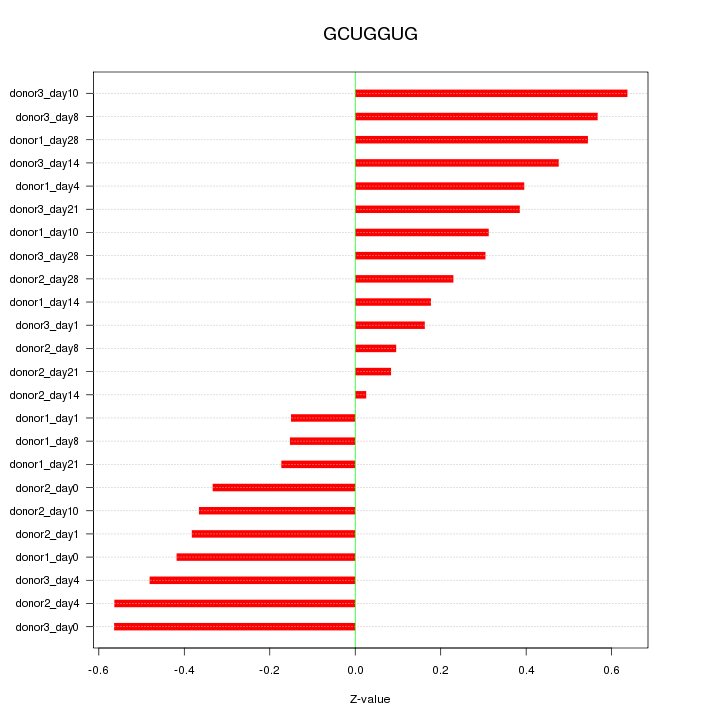
| 0.2 |
0.6 |
GO:1901860 |
positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.1 |
0.4 |
GO:0097477 |
lateral motor column neuron migration(GO:0097477) |
| 0.1 |
0.3 |
GO:2000364 |
regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.1 |
0.1 |
GO:0071335 |
fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) submandibular salivary gland formation(GO:0060661) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.1 |
0.2 |
GO:0015993 |
L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 |
0.5 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 |
0.2 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 |
0.1 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.0 |
0.1 |
GO:0051097 |
negative regulation of helicase activity(GO:0051097) |
| 0.0 |
0.1 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.0 |
0.1 |
GO:0071879 |
positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 |
0.8 |
GO:0032012 |
regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.1 |
GO:0042264 |
peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 |
0.1 |
GO:1901675 |
negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 |
0.3 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.0 |
0.4 |
GO:1990416 |
cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 |
0.1 |
GO:0032484 |
Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 |
0.1 |
GO:1990926 |
short-term synaptic potentiation(GO:1990926) |
| 0.0 |
0.2 |
GO:0060235 |
lens induction in camera-type eye(GO:0060235) |
| 0.0 |
0.3 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.0 |
0.2 |
GO:0007042 |
lysosomal lumen acidification(GO:0007042) |
| 0.0 |
0.1 |
GO:0042713 |
sperm ejaculation(GO:0042713) |
| 0.0 |
0.1 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.0 |
0.1 |
GO:0019722 |
calcium-mediated signaling(GO:0019722) |
| 0.0 |
0.3 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.1 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.0 |
0.1 |
GO:0021823 |
cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 |
0.1 |
GO:0019918 |
peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 |
0.1 |
GO:0030950 |
establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 |
0.2 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 |
0.1 |
GO:0034670 |
chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 |
0.1 |
GO:0008063 |
Toll signaling pathway(GO:0008063) |
| 0.0 |
0.2 |
GO:0031087 |
deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 |
0.1 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.0 |
0.2 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.2 |
GO:0042699 |
follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 |
0.1 |
GO:0061687 |
detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 |
0.0 |
GO:0002101 |
tRNA wobble cytosine modification(GO:0002101) |
| 0.0 |
0.1 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.0 |
0.0 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 |
0.0 |
GO:0031587 |
positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 |
0.0 |
GO:1902956 |
regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.0 |
0.1 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 |
0.0 |
GO:0060448 |
dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 |
0.1 |
GO:0038203 |
TORC2 signaling(GO:0038203) |
| 0.0 |
0.2 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.2 |
GO:0060033 |
anatomical structure regression(GO:0060033) |