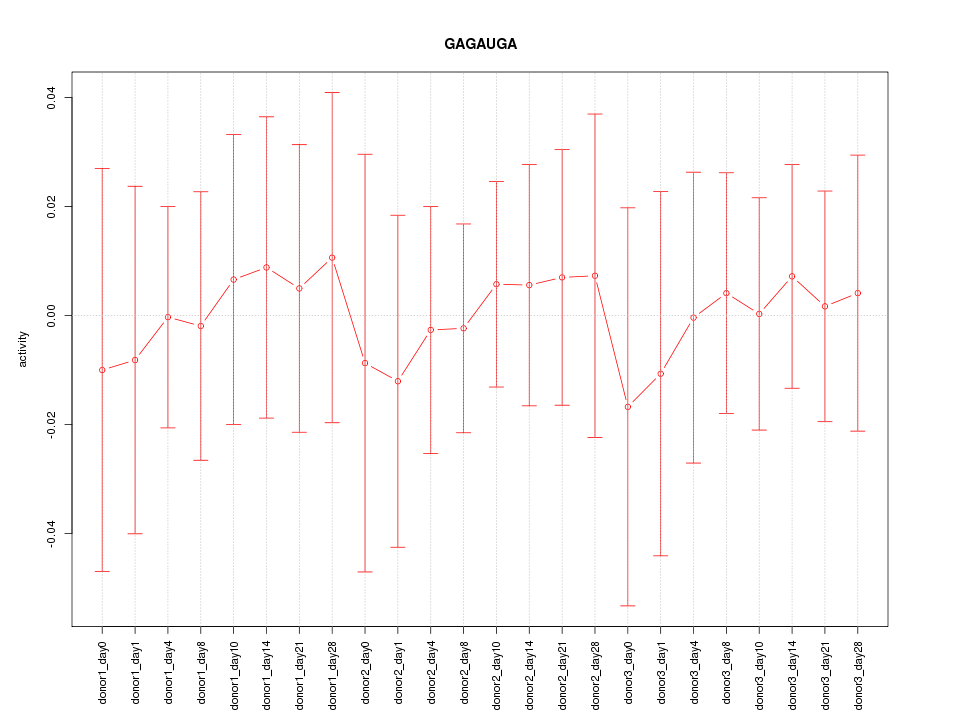
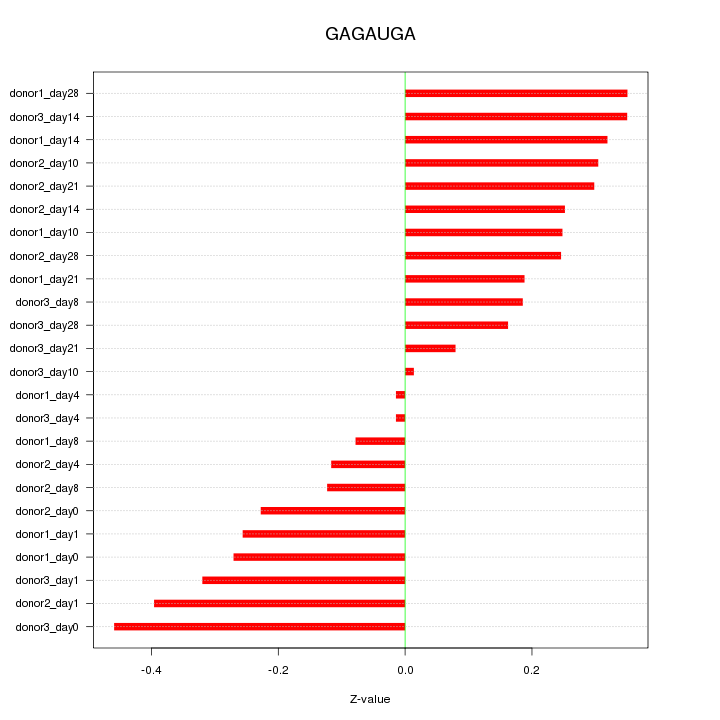
| 0.2 |
0.5 |
GO:0034059 |
response to anoxia(GO:0034059) |
| 0.1 |
0.4 |
GO:1904204 |
regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.1 |
0.4 |
GO:0033634 |
positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 |
0.2 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 |
0.2 |
GO:0035669 |
TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.1 |
0.8 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.2 |
GO:0046909 |
intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 |
0.2 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 |
0.5 |
GO:0042659 |
regulation of cell fate specification(GO:0042659) |
| 0.0 |
0.1 |
GO:0072720 |
response to dithiothreitol(GO:0072720) |
| 0.0 |
0.1 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) positive regulation of NK T cell differentiation(GO:0051138) germline stem cell asymmetric division(GO:0098728) |
| 0.0 |
0.1 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) |
| 0.0 |
0.1 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.0 |
0.2 |
GO:0042985 |
negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 |
0.1 |
GO:0000294 |
nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 |
0.1 |
GO:0048861 |
leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 |
0.2 |
GO:2000467 |
positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 |
0.1 |
GO:0060490 |
orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 |
0.0 |
GO:0021896 |
forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.0 |
0.1 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 |
0.2 |
GO:0001514 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.0 |
GO:0019072 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 |
0.1 |
GO:0001555 |
oocyte growth(GO:0001555) |
| 0.0 |
0.1 |
GO:0060013 |
righting reflex(GO:0060013) |
| 0.0 |
0.1 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.0 |
0.0 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.0 |
0.0 |
GO:0050823 |
peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.0 |
0.9 |
GO:0010830 |
regulation of myotube differentiation(GO:0010830) |
| 0.0 |
0.0 |
GO:0046619 |
optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 |
0.2 |
GO:0042340 |
keratan sulfate catabolic process(GO:0042340) |