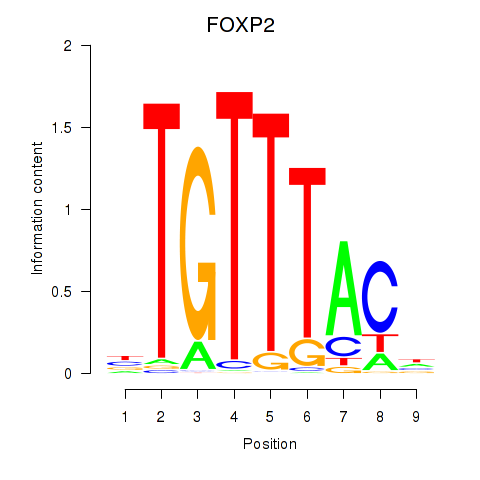
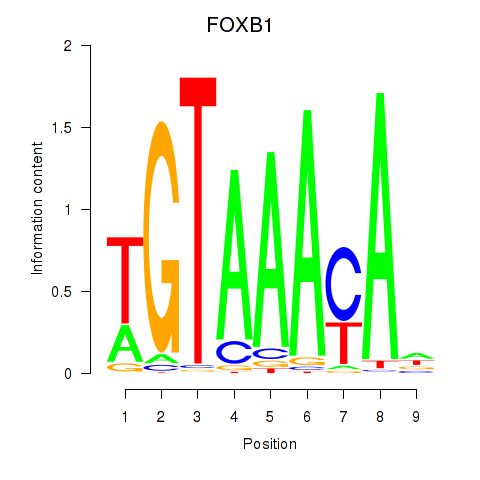
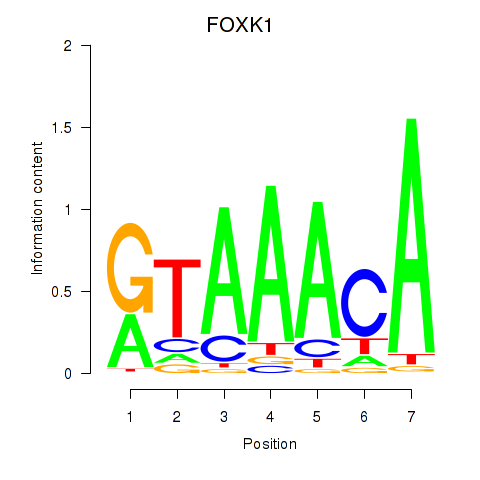
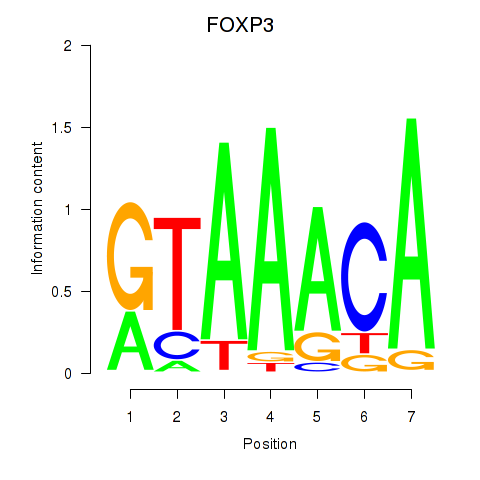
| 1.2 |
3.5 |
GO:0010430 |
fatty acid omega-oxidation(GO:0010430) |
| 1.0 |
3.0 |
GO:1904899 |
regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.9 |
6.9 |
GO:0009441 |
glycolate metabolic process(GO:0009441) |
| 0.5 |
1.6 |
GO:0002071 |
glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.5 |
3.4 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.3 |
0.8 |
GO:0045957 |
regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.2 |
1.5 |
GO:1990822 |
basic amino acid transmembrane transport(GO:1990822) |
| 0.2 |
0.7 |
GO:0098582 |
innate vocalization behavior(GO:0098582) |
| 0.2 |
0.7 |
GO:0030505 |
inorganic diphosphate transport(GO:0030505) |
| 0.2 |
3.6 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.2 |
0.8 |
GO:0072134 |
hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme morphogenesis(GO:0072134) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.2 |
0.6 |
GO:1902725 |
negative regulation of satellite cell differentiation(GO:1902725) |
| 0.2 |
0.2 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.2 |
1.1 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.2 |
0.5 |
GO:0060298 |
positive regulation of sarcomere organization(GO:0060298) |
| 0.2 |
0.4 |
GO:0045349 |
interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.2 |
1.2 |
GO:0044245 |
polysaccharide digestion(GO:0044245) |
| 0.2 |
1.0 |
GO:0006651 |
diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 |
0.5 |
GO:0002514 |
B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.2 |
0.5 |
GO:0070173 |
regulation of enamel mineralization(GO:0070173) |
| 0.2 |
0.5 |
GO:0019521 |
aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.2 |
0.6 |
GO:0035720 |
intraciliary anterograde transport(GO:0035720) |
| 0.2 |
0.9 |
GO:2001034 |
positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.2 |
0.5 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 |
0.3 |
GO:0001970 |
positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.1 |
0.4 |
GO:0042495 |
detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 |
1.7 |
GO:0045078 |
positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 |
1.2 |
GO:0002318 |
myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 |
0.6 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 |
0.4 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 |
1.3 |
GO:0045329 |
carnitine biosynthetic process(GO:0045329) |
| 0.1 |
0.4 |
GO:1990926 |
short-term synaptic potentiation(GO:1990926) |
| 0.1 |
1.0 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.1 |
0.2 |
GO:0061054 |
dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.1 |
0.6 |
GO:0003051 |
angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 |
0.6 |
GO:0061055 |
myotome development(GO:0061055) |
| 0.1 |
0.3 |
GO:0035604 |
fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.1 |
4.3 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.1 |
0.6 |
GO:0044336 |
canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 |
0.3 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 |
1.5 |
GO:0042738 |
exogenous drug catabolic process(GO:0042738) |
| 0.1 |
0.6 |
GO:0008063 |
Toll signaling pathway(GO:0008063) |
| 0.1 |
1.4 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.1 |
0.9 |
GO:1902856 |
negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.1 |
0.6 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 |
0.6 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 0.1 |
0.3 |
GO:0071557 |
negative regulation of chromatin silencing at rDNA(GO:0061188) histone H3-K27 demethylation(GO:0071557) |
| 0.1 |
0.5 |
GO:0051029 |
rRNA transport(GO:0051029) |
| 0.1 |
0.5 |
GO:0044861 |
protein transport into plasma membrane raft(GO:0044861) |
| 0.1 |
0.6 |
GO:1902261 |
positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 |
0.5 |
GO:0010520 |
regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 |
1.0 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.1 |
0.2 |
GO:0014016 |
neuroblast differentiation(GO:0014016) |
| 0.1 |
0.8 |
GO:0045602 |
negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 |
2.1 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.1 |
0.7 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 |
0.3 |
GO:1900169 |
regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 |
2.0 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
0.3 |
GO:0001505 |
regulation of neurotransmitter levels(GO:0001505) |
| 0.1 |
2.5 |
GO:0060972 |
left/right pattern formation(GO:0060972) |
| 0.1 |
0.3 |
GO:0042704 |
uterine wall breakdown(GO:0042704) |
| 0.1 |
0.9 |
GO:0031087 |
deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 |
0.3 |
GO:0043335 |
protein unfolding(GO:0043335) |
| 0.1 |
1.1 |
GO:0046600 |
negative regulation of centriole replication(GO:0046600) |
| 0.1 |
0.3 |
GO:2000502 |
negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 |
0.4 |
GO:1902617 |
response to fluoride(GO:1902617) |
| 0.1 |
0.2 |
GO:0061394 |
regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 |
0.4 |
GO:0043029 |
T cell homeostasis(GO:0043029) |
| 0.1 |
6.7 |
GO:1900047 |
negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 |
0.1 |
GO:1903487 |
regulation of lactation(GO:1903487) |
| 0.1 |
0.2 |
GO:0002933 |
lipid hydroxylation(GO:0002933) |
| 0.1 |
0.3 |
GO:0086097 |
phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 |
0.3 |
GO:0006391 |
transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 |
0.6 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.1 |
0.2 |
GO:0015722 |
canalicular bile acid transport(GO:0015722) |
| 0.1 |
0.2 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 |
0.3 |
GO:0061358 |
negative regulation of Wnt protein secretion(GO:0061358) |
| 0.1 |
0.2 |
GO:0036023 |
limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.1 |
0.3 |
GO:0070997 |
neuron death(GO:0070997) |
| 0.1 |
0.4 |
GO:0002669 |
positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 |
0.2 |
GO:0051835 |
positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 |
0.1 |
GO:0002581 |
negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 |
0.4 |
GO:0045074 |
interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 |
1.2 |
GO:0090360 |
platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 |
3.3 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.1 |
0.1 |
GO:0052314 |
phytoalexin metabolic process(GO:0052314) |
| 0.1 |
0.3 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 |
0.6 |
GO:0001714 |
endodermal cell fate specification(GO:0001714) |
| 0.1 |
0.3 |
GO:0000255 |
allantoin metabolic process(GO:0000255) |
| 0.1 |
0.2 |
GO:0021986 |
epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 |
0.2 |
GO:0002728 |
negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 |
0.4 |
GO:1903385 |
regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 |
0.3 |
GO:0035860 |
glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 |
1.2 |
GO:0019373 |
epoxygenase P450 pathway(GO:0019373) |
| 0.1 |
0.3 |
GO:0060018 |
astrocyte fate commitment(GO:0060018) |
| 0.1 |
0.1 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 |
0.4 |
GO:0097056 |
selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 |
0.6 |
GO:0021520 |
spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 |
0.1 |
GO:0060738 |
epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.1 |
0.2 |
GO:0034059 |
response to anoxia(GO:0034059) |
| 0.1 |
1.1 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 |
0.2 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) fasciculation of motor neuron axon(GO:0097156) |
| 0.1 |
0.2 |
GO:0051933 |
amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 |
0.5 |
GO:0046604 |
positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 |
0.2 |
GO:0097069 |
cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.1 |
0.4 |
GO:0071918 |
urea transmembrane transport(GO:0071918) |
| 0.1 |
0.6 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.1 |
0.3 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.1 |
0.2 |
GO:0090299 |
regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 |
0.2 |
GO:2000547 |
regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.1 |
0.6 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.1 |
1.3 |
GO:0072189 |
ureter development(GO:0072189) |
| 0.1 |
1.0 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 |
0.8 |
GO:0035372 |
protein localization to microtubule(GO:0035372) |
| 0.1 |
0.2 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 |
0.2 |
GO:0042214 |
terpene metabolic process(GO:0042214) |
| 0.1 |
0.6 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 |
0.3 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 |
1.1 |
GO:0021670 |
lateral ventricle development(GO:0021670) |
| 0.1 |
0.4 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 |
0.2 |
GO:0038043 |
interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 |
0.2 |
GO:0093001 |
glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.1 |
0.3 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 |
0.2 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.1 |
0.5 |
GO:0016191 |
synaptic vesicle uncoating(GO:0016191) |
| 0.1 |
0.4 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.1 |
0.1 |
GO:0035711 |
T-helper 1 cell activation(GO:0035711) |
| 0.1 |
0.2 |
GO:0042737 |
drug catabolic process(GO:0042737) |
| 0.1 |
0.3 |
GO:0006196 |
AMP catabolic process(GO:0006196) |
| 0.1 |
0.1 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.2 |
GO:0034392 |
negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 |
0.1 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.2 |
GO:0032474 |
otolith morphogenesis(GO:0032474) |
| 0.0 |
0.1 |
GO:0072106 |
regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.0 |
0.2 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 |
0.2 |
GO:0019087 |
transformation of host cell by virus(GO:0019087) |
| 0.0 |
0.2 |
GO:0046782 |
regulation of viral transcription(GO:0046782) |
| 0.0 |
0.2 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 |
0.1 |
GO:0070839 |
divalent metal ion export(GO:0070839) |
| 0.0 |
0.1 |
GO:0035994 |
response to muscle stretch(GO:0035994) |
| 0.0 |
0.5 |
GO:0021562 |
vestibulocochlear nerve development(GO:0021562) |
| 0.0 |
0.2 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.0 |
0.2 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.0 |
0.3 |
GO:0006499 |
N-terminal protein myristoylation(GO:0006499) |
| 0.0 |
0.3 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.4 |
GO:1903802 |
L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.0 |
0.2 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 |
0.4 |
GO:1900227 |
positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 |
0.2 |
GO:0009956 |
radial pattern formation(GO:0009956) |
| 0.0 |
0.5 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.0 |
0.2 |
GO:0019556 |
histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 |
0.2 |
GO:0048698 |
negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 |
0.5 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 |
1.0 |
GO:1904886 |
beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 |
0.1 |
GO:0033603 |
positive regulation of dopamine secretion(GO:0033603) |
| 0.0 |
0.5 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 |
0.2 |
GO:0033591 |
response to L-ascorbic acid(GO:0033591) |
| 0.0 |
0.2 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 |
3.0 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.0 |
0.2 |
GO:0044026 |
DNA hypermethylation(GO:0044026) |
| 0.0 |
0.3 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 |
0.3 |
GO:0003417 |
growth plate cartilage development(GO:0003417) |
| 0.0 |
0.3 |
GO:1901385 |
regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 |
0.1 |
GO:0018032 |
protein amidation(GO:0018032) |
| 0.0 |
0.1 |
GO:0010387 |
COP9 signalosome assembly(GO:0010387) |
| 0.0 |
0.2 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 |
0.4 |
GO:0031344 |
regulation of cell projection organization(GO:0031344) |
| 0.0 |
0.3 |
GO:0021514 |
ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 |
0.1 |
GO:0001545 |
primary ovarian follicle growth(GO:0001545) |
| 0.0 |
0.1 |
GO:0035674 |
tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 |
0.1 |
GO:0002418 |
immune response to tumor cell(GO:0002418) |
| 0.0 |
0.3 |
GO:0032277 |
negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 |
0.3 |
GO:0042996 |
regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 |
0.1 |
GO:0006533 |
aspartate catabolic process(GO:0006533) |
| 0.0 |
0.1 |
GO:0051958 |
methotrexate transport(GO:0051958) |
| 0.0 |
0.1 |
GO:0036496 |
regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 |
0.3 |
GO:0061669 |
spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 |
0.3 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.0 |
0.1 |
GO:0006001 |
fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 |
0.1 |
GO:0035026 |
leading edge cell differentiation(GO:0035026) |
| 0.0 |
0.6 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 |
1.1 |
GO:0043161 |
proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 |
0.1 |
GO:0031860 |
telomeric 3' overhang formation(GO:0031860) |
| 0.0 |
0.3 |
GO:1903546 |
protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 |
0.1 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.0 |
0.1 |
GO:1903215 |
negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 |
0.2 |
GO:0045959 |
regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 |
0.4 |
GO:0044130 |
negative regulation of growth of symbiont in host(GO:0044130) |
| 0.0 |
0.1 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 |
0.1 |
GO:0006864 |
pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 |
0.2 |
GO:1902748 |
melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 |
0.3 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 |
0.5 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.0 |
1.3 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.2 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.0 |
0.3 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.0 |
0.1 |
GO:0060300 |
regulation of cytokine activity(GO:0060300) |
| 0.0 |
2.3 |
GO:0001895 |
retina homeostasis(GO:0001895) |
| 0.0 |
0.0 |
GO:0032661 |
regulation of interleukin-18 production(GO:0032661) |
| 0.0 |
0.1 |
GO:0002904 |
positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 |
0.4 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 |
0.1 |
GO:0015842 |
aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 |
0.1 |
GO:0006990 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 |
0.1 |
GO:0006617 |
SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 |
0.3 |
GO:0006987 |
activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 |
1.3 |
GO:0050690 |
regulation of defense response to virus by virus(GO:0050690) |
| 0.0 |
0.1 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.0 |
0.1 |
GO:0045964 |
positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 |
0.2 |
GO:0051388 |
positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 |
0.1 |
GO:0044240 |
multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 |
0.2 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.0 |
0.2 |
GO:0042780 |
tRNA 3'-end processing(GO:0042780) |
| 0.0 |
0.1 |
GO:0035936 |
testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 |
1.2 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.2 |
GO:0000066 |
mitochondrial ornithine transport(GO:0000066) |
| 0.0 |
0.3 |
GO:0006069 |
ethanol oxidation(GO:0006069) |
| 0.0 |
0.1 |
GO:0006579 |
amino-acid betaine catabolic process(GO:0006579) |
| 0.0 |
0.1 |
GO:0010902 |
positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.0 |
0.1 |
GO:0035995 |
detection of muscle stretch(GO:0035995) |
| 0.0 |
0.1 |
GO:0051182 |
coenzyme transport(GO:0051182) |
| 0.0 |
0.1 |
GO:0051083 |
'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 |
0.1 |
GO:0006114 |
glycerol biosynthetic process(GO:0006114) |
| 0.0 |
0.7 |
GO:0070734 |
histone H3-K27 methylation(GO:0070734) |
| 0.0 |
0.2 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.0 |
0.3 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 |
0.2 |
GO:0036500 |
ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 |
0.4 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.0 |
GO:1902956 |
regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.0 |
0.2 |
GO:0030885 |
regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 |
0.2 |
GO:0035457 |
cellular response to interferon-alpha(GO:0035457) |
| 0.0 |
0.1 |
GO:1902741 |
interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 |
0.1 |
GO:0070127 |
tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 |
0.2 |
GO:2000741 |
asymmetric neuroblast division(GO:0055059) positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 |
0.2 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.1 |
GO:0099540 |
synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 |
0.1 |
GO:1902998 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 |
0.2 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 |
0.2 |
GO:0097428 |
protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 |
0.1 |
GO:0039663 |
fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 |
0.1 |
GO:0050773 |
regulation of dendrite development(GO:0050773) |
| 0.0 |
0.8 |
GO:2000249 |
regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 |
0.2 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 |
0.1 |
GO:0045425 |
positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 |
0.0 |
GO:0043486 |
histone exchange(GO:0043486) |
| 0.0 |
0.2 |
GO:0006552 |
leucine catabolic process(GO:0006552) |
| 0.0 |
0.2 |
GO:0090204 |
protein localization to nuclear pore(GO:0090204) |
| 0.0 |
0.1 |
GO:0016260 |
selenocysteine metabolic process(GO:0016259) selenocysteine biosynthetic process(GO:0016260) |
| 0.0 |
0.0 |
GO:0060214 |
endocardium formation(GO:0060214) |
| 0.0 |
0.1 |
GO:0008627 |
intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 |
0.1 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 |
0.1 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 |
0.2 |
GO:0042428 |
serotonin metabolic process(GO:0042428) |
| 0.0 |
0.7 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.0 |
0.2 |
GO:1903800 |
positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 |
0.1 |
GO:0002224 |
pattern recognition receptor signaling pathway(GO:0002221) toll-like receptor signaling pathway(GO:0002224) |
| 0.0 |
0.0 |
GO:0048021 |
regulation of melanin biosynthetic process(GO:0048021) positive regulation of melanin biosynthetic process(GO:0048023) regulation of secondary metabolite biosynthetic process(GO:1900376) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 |
0.0 |
GO:0042035 |
regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 |
0.1 |
GO:0051270 |
regulation of cellular component movement(GO:0051270) |
| 0.0 |
0.4 |
GO:0019614 |
catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 |
0.1 |
GO:0010949 |
negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 0.0 |
0.2 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.2 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 |
0.2 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.0 |
0.1 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.0 |
0.1 |
GO:0043570 |
maintenance of DNA repeat elements(GO:0043570) |
| 0.0 |
0.1 |
GO:1903644 |
regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 |
0.1 |
GO:0070781 |
response to biotin(GO:0070781) |
| 0.0 |
0.2 |
GO:0072383 |
plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 |
0.3 |
GO:0070307 |
lens fiber cell development(GO:0070307) |
| 0.0 |
0.7 |
GO:0045745 |
positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 |
0.0 |
GO:0052151 |
positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 |
0.2 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.1 |
GO:0032764 |
negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 |
0.1 |
GO:0090191 |
negative regulation of mesonephros development(GO:0061218) negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.0 |
0.2 |
GO:0014820 |
tonic smooth muscle contraction(GO:0014820) |
| 0.0 |
0.1 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) |
| 0.0 |
0.4 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.0 |
0.3 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.0 |
0.1 |
GO:0043324 |
eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 |
0.1 |
GO:1903906 |
plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 |
0.1 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.0 |
0.1 |
GO:0052055 |
modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 |
0.2 |
GO:0061088 |
regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 |
0.1 |
GO:0070528 |
protein kinase C signaling(GO:0070528) |
| 0.0 |
3.2 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.5 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.0 |
0.1 |
GO:0051051 |
negative regulation of transport(GO:0051051) |
| 0.0 |
0.2 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 |
0.1 |
GO:1902412 |
regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 |
0.2 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.0 |
2.1 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
1.2 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 |
0.1 |
GO:0071349 |
interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.0 |
0.1 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 |
0.1 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 |
0.1 |
GO:0030048 |
actin filament-based movement(GO:0030048) |
| 0.0 |
0.4 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.0 |
0.1 |
GO:0015783 |
GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 |
0.3 |
GO:2000394 |
positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 |
0.1 |
GO:0071360 |
cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 |
0.0 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 |
0.1 |
GO:0038161 |
prolactin signaling pathway(GO:0038161) |
| 0.0 |
0.1 |
GO:0008218 |
bioluminescence(GO:0008218) |
| 0.0 |
0.1 |
GO:0061760 |
antifungal innate immune response(GO:0061760) |
| 0.0 |
0.0 |
GO:0045900 |
negative regulation of translational elongation(GO:0045900) |
| 0.0 |
0.3 |
GO:0050930 |
induction of positive chemotaxis(GO:0050930) |
| 0.0 |
0.1 |
GO:0003351 |
epithelial cilium movement(GO:0003351) |
| 0.0 |
0.1 |
GO:1900020 |
regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 |
0.1 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.1 |
GO:0032627 |
interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 |
0.1 |
GO:0002088 |
lens development in camera-type eye(GO:0002088) |
| 0.0 |
0.3 |
GO:0032354 |
response to follicle-stimulating hormone(GO:0032354) |
| 0.0 |
0.1 |
GO:0003172 |
primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) regulation of synaptic activity(GO:0060025) |
| 0.0 |
0.1 |
GO:0042126 |
nitrate metabolic process(GO:0042126) |
| 0.0 |
0.2 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.0 |
0.1 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 |
0.1 |
GO:0050999 |
regulation of monooxygenase activity(GO:0032768) regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.0 |
0.1 |
GO:0090343 |
positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.0 |
0.0 |
GO:2000395 |
regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 |
0.1 |
GO:0070172 |
positive regulation of tooth mineralization(GO:0070172) |
| 0.0 |
0.0 |
GO:0061428 |
negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 |
0.1 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.0 |
0.1 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.2 |
GO:0044849 |
estrous cycle(GO:0044849) |
| 0.0 |
1.1 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.1 |
GO:0048010 |
vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 |
0.3 |
GO:0036150 |
phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 |
1.3 |
GO:0097031 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.2 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.0 |
0.1 |
GO:0034285 |
response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 |
0.1 |
GO:1904781 |
positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 |
0.2 |
GO:0043116 |
negative regulation of vascular permeability(GO:0043116) |
| 0.0 |
0.1 |
GO:0006848 |
pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 |
0.0 |
GO:0006286 |
base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 |
0.0 |
GO:0090669 |
telomerase RNA stabilization(GO:0090669) |
| 0.0 |
0.3 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 |
0.1 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.0 |
0.1 |
GO:1902897 |
regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 |
0.1 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.1 |
GO:0050916 |
sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 |
0.0 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.0 |
0.1 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 |
0.1 |
GO:0033387 |
putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 |
0.2 |
GO:0035810 |
positive regulation of urine volume(GO:0035810) |
| 0.0 |
0.1 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 |
0.1 |
GO:0060770 |
negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 |
0.2 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 |
0.1 |
GO:0030490 |
maturation of SSU-rRNA(GO:0030490) |
| 0.0 |
0.2 |
GO:0006913 |
nucleocytoplasmic transport(GO:0006913) |
| 0.0 |
0.5 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
0.1 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.0 |
0.1 |
GO:0060088 |
auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 |
0.1 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.1 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.1 |
GO:0038155 |
interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 |
0.7 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
0.1 |
GO:0051122 |
hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 |
0.3 |
GO:0046856 |
phospholipid dephosphorylation(GO:0046839) phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 |
0.4 |
GO:0046326 |
positive regulation of glucose import(GO:0046326) |
| 0.0 |
0.0 |
GO:0007606 |
sensory perception of chemical stimulus(GO:0007606) |
| 0.0 |
0.1 |
GO:1903896 |
positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.0 |
0.1 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 |
0.0 |
GO:0003139 |
secondary heart field specification(GO:0003139) |
| 0.0 |
0.1 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.0 |
0.1 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
0.2 |
GO:0070129 |
regulation of mitochondrial translation(GO:0070129) |
| 0.0 |
0.1 |
GO:0015781 |
nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 |
0.1 |
GO:0060737 |
prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.0 |
0.0 |
GO:0019417 |
sulfur oxidation(GO:0019417) |
| 0.0 |
0.0 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 |
0.1 |
GO:0002084 |
protein depalmitoylation(GO:0002084) |
| 0.0 |
0.2 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.0 |
0.0 |
GO:2000035 |
regulation of stem cell division(GO:2000035) |
| 0.0 |
0.5 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.1 |
GO:0001711 |
endodermal cell fate commitment(GO:0001711) |
| 0.0 |
0.4 |
GO:0030318 |
melanocyte differentiation(GO:0030318) |
| 0.0 |
0.1 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.1 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.0 |
0.1 |
GO:0006121 |
mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 |
0.0 |
GO:0042701 |
progesterone secretion(GO:0042701) |
| 0.0 |
0.4 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 |
0.0 |
GO:2000074 |
regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 |
0.1 |
GO:1900103 |
positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 |
0.2 |
GO:0040015 |
negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 |
0.2 |
GO:1904355 |
positive regulation of telomere capping(GO:1904355) |
| 0.0 |
0.2 |
GO:0035640 |
exploration behavior(GO:0035640) |
| 0.0 |
0.1 |
GO:0097105 |
presynaptic membrane assembly(GO:0097105) |
| 0.0 |
0.1 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 |
0.1 |
GO:0070301 |
cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 |
0.1 |
GO:0016559 |
peroxisome fission(GO:0016559) |
| 0.0 |
0.4 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.3 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.0 |
0.0 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.0 |
0.2 |
GO:0036315 |
cellular response to sterol(GO:0036315) |
| 0.0 |
0.0 |
GO:0060492 |
foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 |
0.1 |
GO:0001712 |
ectodermal cell fate commitment(GO:0001712) |
| 0.0 |
0.3 |
GO:0090200 |
positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 |
0.1 |
GO:2000042 |
negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 |
0.0 |
GO:0006711 |
estrogen catabolic process(GO:0006711) |