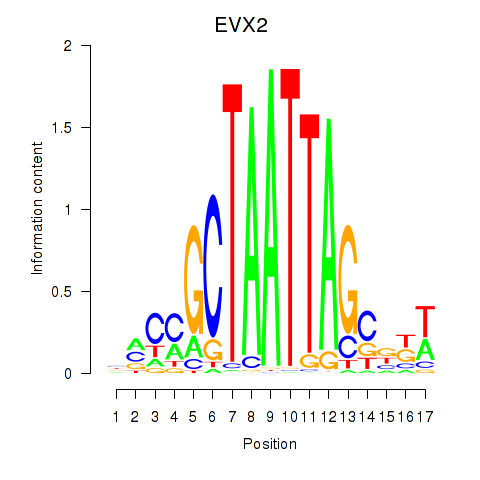
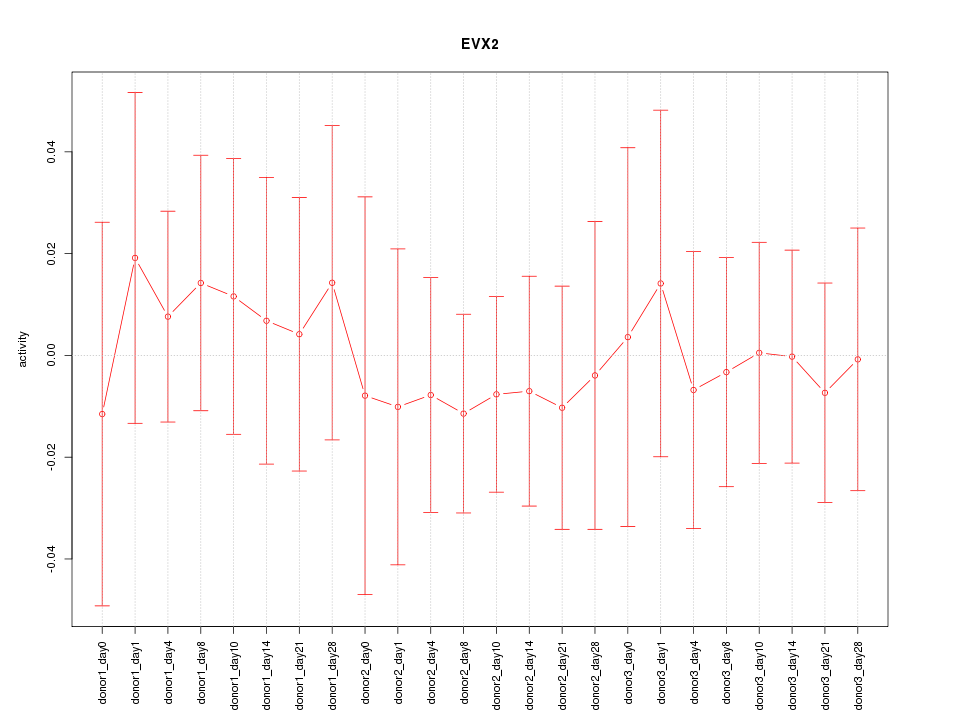
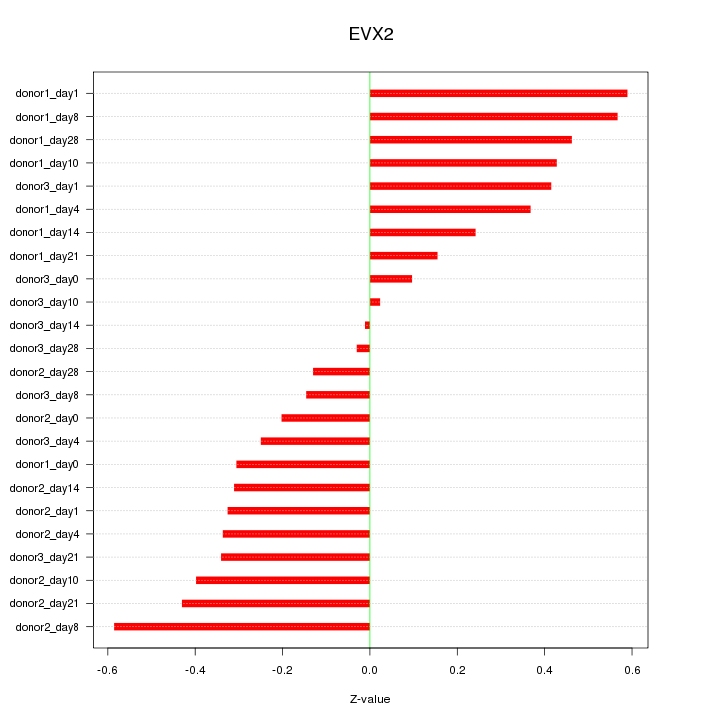
|
chr3_+_111718173
|
0.816
|
ENST00000494932.1
|
TAGLN3
|
transgelin 3
|
|
chr11_-_115375107
|
0.802
|
ENST00000545380.1
ENST00000452722.3
ENST00000537058.1
ENST00000536727.1
ENST00000542447.2
ENST00000331581.6
|
CADM1
|
cell adhesion molecule 1
|
|
chr3_+_111717600
|
0.690
|
ENST00000273368.4
|
TAGLN3
|
transgelin 3
|
|
chr3_+_111718036
|
0.668
|
ENST00000455401.2
|
TAGLN3
|
transgelin 3
|
|
chr3_+_111717511
|
0.646
|
ENST00000478951.1
ENST00000393917.2
|
TAGLN3
|
transgelin 3
|
|
chr20_-_50418972
|
0.355
|
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4
|
|
chr20_-_50418947
|
0.336
|
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4
|
|
chr20_-_50419055
|
0.273
|
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4
|
|
chr20_+_43990576
|
0.216
|
ENST00000372727.1
ENST00000414310.1
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr13_+_78315466
|
0.203
|
ENST00000314070.5
ENST00000462234.1
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr1_+_28261492
|
0.196
|
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B
|
|
chr1_+_28261621
|
0.177
|
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B
|
|
chr4_+_144354644
|
0.174
|
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1
|
|
chr4_-_41884620
|
0.149
|
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682
|
|
chr5_-_96478466
|
0.147
|
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken)
|
|
chr4_-_41884582
|
0.122
|
ENST00000499082.2
|
LINC00682
|
long intergenic non-protein coding RNA 682
|
|
chr1_-_151431647
|
0.104
|
ENST00000368863.2
ENST00000409503.1
ENST00000491586.1
ENST00000533351.1
ENST00000540984.1
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr6_+_39760129
|
0.080
|
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr6_-_62996066
|
0.076
|
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2
|
|
chr1_+_28261533
|
0.073
|
ENST00000411604.1
ENST00000373888.4
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B
|
|
chr1_-_151431909
|
0.054
|
ENST00000361398.3
ENST00000271715.2
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr12_+_28410128
|
0.040
|
ENST00000381259.1
ENST00000381256.1
|
CCDC91
|
coiled-coil domain containing 91
|
|
chr17_+_18086392
|
0.032
|
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli)
|
|
chr17_-_9929581
|
0.022
|
ENST00000437099.2
ENST00000396115.2
|
GAS7
|
growth arrest-specific 7
|
|
chr6_-_111927062
|
0.009
|
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2
|