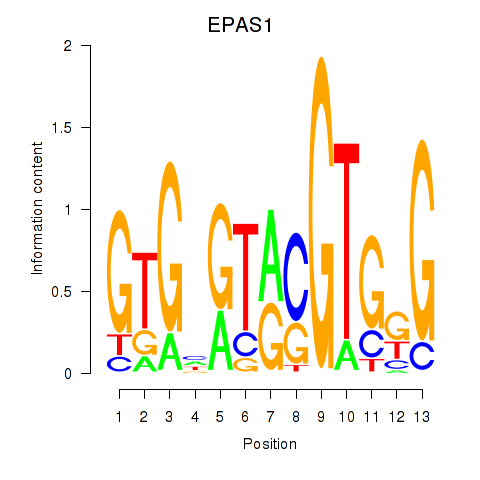
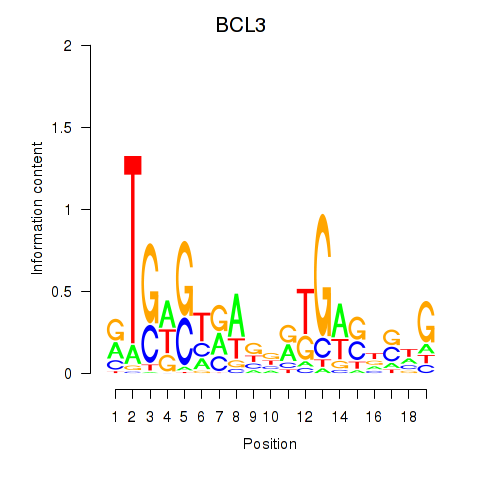
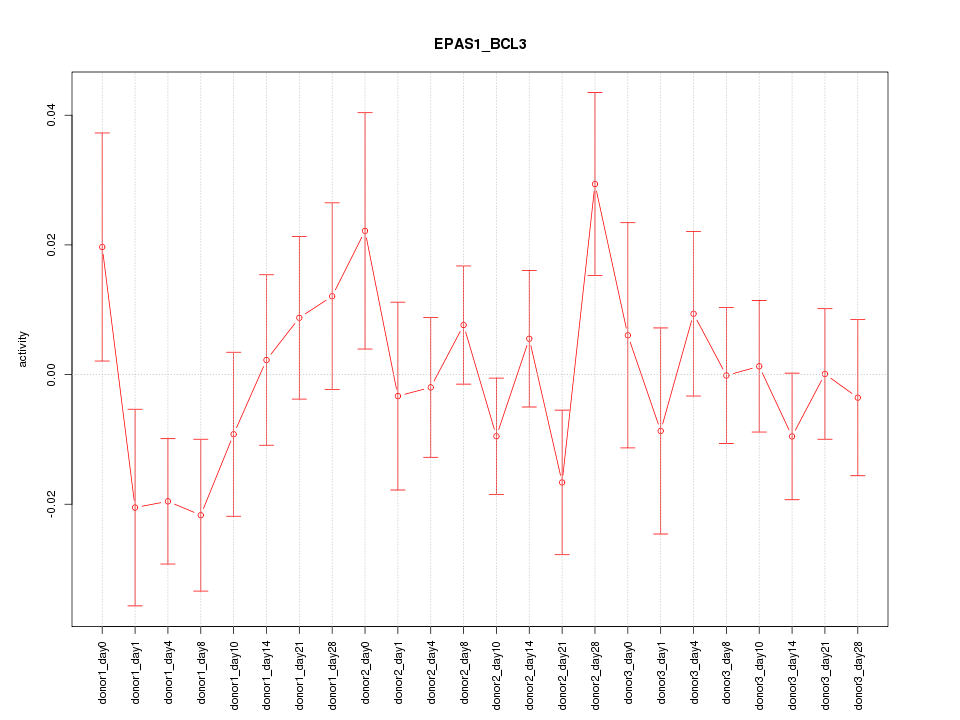
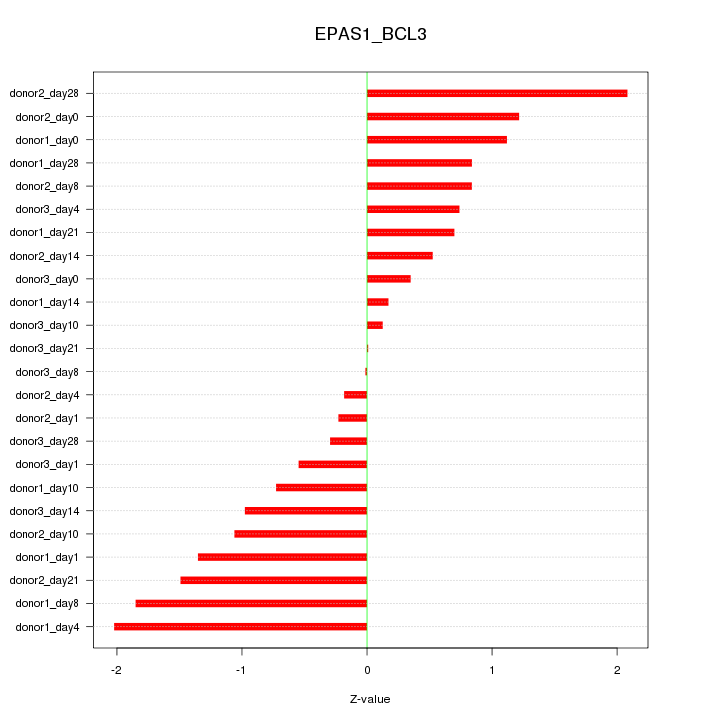
| 0.3 |
0.9 |
GO:1904328 |
regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.3 |
1.0 |
GO:0046271 |
phenylpropanoid catabolic process(GO:0046271) |
| 0.2 |
0.7 |
GO:0061537 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.2 |
0.8 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 |
1.5 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 |
1.1 |
GO:0010734 |
protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.2 |
0.5 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.2 |
0.2 |
GO:0099545 |
trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.2 |
1.4 |
GO:0021564 |
vagus nerve development(GO:0021564) |
| 0.2 |
0.5 |
GO:0002023 |
reduction of food intake in response to dietary excess(GO:0002023) |
| 0.2 |
0.5 |
GO:0061054 |
dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.1 |
1.0 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.1 |
0.4 |
GO:0018874 |
benzoate metabolic process(GO:0018874) |
| 0.1 |
0.5 |
GO:1901842 |
negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 |
0.3 |
GO:0072034 |
renal vesicle induction(GO:0072034) |
| 0.1 |
0.5 |
GO:0072299 |
negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.1 |
0.4 |
GO:0010621 |
negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.1 |
0.6 |
GO:0044208 |
'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 |
0.6 |
GO:1901731 |
positive regulation of platelet aggregation(GO:1901731) |
| 0.1 |
0.3 |
GO:0003025 |
regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.1 |
0.3 |
GO:0034445 |
regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.1 |
0.3 |
GO:1904298 |
positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 0.1 |
0.3 |
GO:0001544 |
initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 |
0.2 |
GO:0009183 |
purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.1 |
0.3 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.1 |
0.4 |
GO:0090362 |
positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 |
0.4 |
GO:1904327 |
protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 |
0.6 |
GO:0006196 |
AMP catabolic process(GO:0006196) |
| 0.1 |
0.6 |
GO:0071895 |
odontoblast differentiation(GO:0071895) |
| 0.1 |
0.3 |
GO:0010725 |
regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.1 |
0.3 |
GO:1903697 |
negative regulation of microvillus assembly(GO:1903697) |
| 0.1 |
0.4 |
GO:0072658 |
maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 |
0.3 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 |
0.2 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 |
0.2 |
GO:0099543 |
positive regulation of adrenergic receptor signaling pathway(GO:0071879) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.1 |
0.4 |
GO:0010961 |
cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 |
0.4 |
GO:0071286 |
cellular response to magnesium ion(GO:0071286) |
| 0.1 |
0.3 |
GO:0070124 |
mitochondrial translational initiation(GO:0070124) |
| 0.1 |
0.2 |
GO:1901876 |
regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 |
0.2 |
GO:2001226 |
negative regulation of chloride transport(GO:2001226) |
| 0.1 |
0.4 |
GO:2000490 |
negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 |
0.9 |
GO:0090240 |
positive regulation of histone H4 acetylation(GO:0090240) |
| 0.1 |
0.2 |
GO:2000364 |
regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.1 |
0.2 |
GO:0050760 |
negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 |
1.4 |
GO:0072540 |
T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 |
0.4 |
GO:0060125 |
negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 |
0.2 |
GO:1990926 |
short-term synaptic potentiation(GO:1990926) |
| 0.1 |
0.3 |
GO:0002803 |
positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 |
0.3 |
GO:0060842 |
arterial endothelial cell differentiation(GO:0060842) |
| 0.1 |
0.1 |
GO:1903526 |
negative regulation of membrane tubulation(GO:1903526) |
| 0.1 |
0.2 |
GO:0051885 |
positive regulation of anagen(GO:0051885) |
| 0.1 |
1.0 |
GO:0035641 |
locomotory exploration behavior(GO:0035641) |
| 0.1 |
0.2 |
GO:0021986 |
epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 |
0.8 |
GO:0007196 |
adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 |
0.2 |
GO:0010816 |
substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 |
0.1 |
GO:0032095 |
regulation of response to food(GO:0032095) |
| 0.1 |
0.4 |
GO:2000323 |
negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 |
0.3 |
GO:0034436 |
glycoprotein transport(GO:0034436) |
| 0.1 |
0.3 |
GO:1901662 |
phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 |
0.5 |
GO:0098914 |
membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 |
0.2 |
GO:2000296 |
negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.1 |
0.2 |
GO:0043006 |
activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.0 |
0.3 |
GO:0070945 |
neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 |
0.3 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.0 |
0.1 |
GO:0031133 |
regulation of axon diameter(GO:0031133) |
| 0.0 |
0.5 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.2 |
GO:1904448 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 |
0.4 |
GO:0031444 |
slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 |
0.6 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 |
0.2 |
GO:0010387 |
COP9 signalosome assembly(GO:0010387) |
| 0.0 |
0.2 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.0 |
0.2 |
GO:0006710 |
androgen catabolic process(GO:0006710) |
| 0.0 |
0.2 |
GO:0038169 |
somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 |
0.3 |
GO:0009957 |
epidermal cell fate specification(GO:0009957) |
| 0.0 |
0.2 |
GO:2000035 |
regulation of stem cell division(GO:2000035) |
| 0.0 |
0.5 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.0 |
0.1 |
GO:0060697 |
positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 |
0.1 |
GO:0008628 |
hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 |
0.1 |
GO:0099624 |
atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.0 |
0.1 |
GO:0036058 |
filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 |
0.1 |
GO:0032278 |
positive regulation of gonadotropin secretion(GO:0032278) |
| 0.0 |
0.1 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.0 |
0.5 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 |
0.2 |
GO:0097411 |
hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 |
0.2 |
GO:0061056 |
sclerotome development(GO:0061056) |
| 0.0 |
0.2 |
GO:0051122 |
hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 |
0.1 |
GO:0021524 |
visceral motor neuron differentiation(GO:0021524) |
| 0.0 |
0.2 |
GO:0070649 |
polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 |
0.1 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 |
0.4 |
GO:0043578 |
nuclear matrix organization(GO:0043578) |
| 0.0 |
0.2 |
GO:1902474 |
positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 |
0.1 |
GO:0044179 |
hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 |
0.1 |
GO:0034473 |
U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 |
0.3 |
GO:0033578 |
protein glycosylation in Golgi(GO:0033578) |
| 0.0 |
0.2 |
GO:0071461 |
cellular response to redox state(GO:0071461) |
| 0.0 |
0.3 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.0 |
0.1 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 |
0.3 |
GO:2000344 |
positive regulation of acrosome reaction(GO:2000344) |
| 0.0 |
1.0 |
GO:0042554 |
superoxide anion generation(GO:0042554) |
| 0.0 |
0.3 |
GO:0090232 |
peripheral nervous system myelin maintenance(GO:0032287) positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 |
0.1 |
GO:1902303 |
regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 |
0.2 |
GO:0060467 |
negative regulation of fertilization(GO:0060467) |
| 0.0 |
0.4 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.2 |
GO:0038110 |
interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 |
0.3 |
GO:0021562 |
vestibulocochlear nerve development(GO:0021562) |
| 0.0 |
0.1 |
GO:0033563 |
dorsal/ventral axon guidance(GO:0033563) |
| 0.0 |
0.3 |
GO:0097011 |
cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 |
0.1 |
GO:2000721 |
positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 |
0.6 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 |
0.1 |
GO:0010949 |
negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 0.0 |
0.2 |
GO:0072675 |
multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 |
0.3 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.0 |
0.2 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 |
0.1 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 |
0.2 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.0 |
0.1 |
GO:0098746 |
fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 |
0.2 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
0.1 |
GO:1902567 |
negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) negative regulation of eosinophil activation(GO:1902567) negative regulation of membrane invagination(GO:1905154) negative regulation of eosinophil migration(GO:2000417) |
| 0.0 |
0.1 |
GO:0043397 |
positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.0 |
0.1 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 |
0.1 |
GO:0072553 |
terminal button organization(GO:0072553) |
| 0.0 |
1.3 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 |
0.2 |
GO:0043117 |
positive regulation of vascular permeability(GO:0043117) |
| 0.0 |
0.1 |
GO:0000711 |
meiotic DNA repair synthesis(GO:0000711) |
| 0.0 |
0.1 |
GO:0099540 |
synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 |
1.2 |
GO:0061718 |
NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 |
0.1 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.0 |
0.0 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 |
0.2 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 |
0.1 |
GO:0097498 |
endothelial tube lumen extension(GO:0097498) |
| 0.0 |
0.2 |
GO:0099638 |
endosome to plasma membrane protein transport(GO:0099638) |
| 0.0 |
0.1 |
GO:1900224 |
positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 |
0.1 |
GO:1990523 |
bone regeneration(GO:1990523) |
| 0.0 |
0.2 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.2 |
GO:0045732 |
positive regulation of protein catabolic process(GO:0045732) |
| 0.0 |
0.0 |
GO:0072717 |
response to actinomycin D(GO:0072716) cellular response to actinomycin D(GO:0072717) |
| 0.0 |
0.1 |
GO:0045586 |
regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.0 |
0.2 |
GO:0018916 |
nitrobenzene metabolic process(GO:0018916) |
| 0.0 |
0.2 |
GO:2000580 |
positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 |
0.1 |
GO:0034154 |
toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 |
0.2 |
GO:0048706 |
embryonic skeletal system development(GO:0048706) |
| 0.0 |
0.1 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.0 |
0.2 |
GO:0032488 |
Cdc42 protein signal transduction(GO:0032488) anterior/posterior axon guidance(GO:0033564) |
| 0.0 |
0.3 |
GO:0090179 |
planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 |
0.1 |
GO:0046878 |
positive regulation of saliva secretion(GO:0046878) |
| 0.0 |
0.1 |
GO:0035502 |
metanephric part of ureteric bud development(GO:0035502) |
| 0.0 |
0.1 |
GO:1903575 |
cornified envelope assembly(GO:1903575) |
| 0.0 |
0.4 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
0.2 |
GO:0030885 |
regulation of myeloid dendritic cell activation(GO:0030885) positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 |
0.2 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 |
0.3 |
GO:0099500 |
synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 |
0.1 |
GO:0002290 |
gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.0 |
0.4 |
GO:0007597 |
blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 |
0.2 |
GO:0003360 |
brainstem development(GO:0003360) |
| 0.0 |
0.1 |
GO:0006554 |
lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 |
0.3 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.2 |
GO:0002329 |
pre-B cell differentiation(GO:0002329) |
| 0.0 |
0.2 |
GO:0010635 |
regulation of mitochondrial fusion(GO:0010635) |
| 0.0 |
0.2 |
GO:0086024 |
adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 |
0.2 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.0 |
0.6 |
GO:1903861 |
positive regulation of dendrite extension(GO:1903861) |
| 0.0 |
0.1 |
GO:0001188 |
RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 |
0.1 |
GO:0070309 |
lens fiber cell morphogenesis(GO:0070309) |
| 0.0 |
0.1 |
GO:0060266 |
negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 |
0.2 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.0 |
0.1 |
GO:1900106 |
hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 |
0.1 |
GO:0061073 |
ciliary body morphogenesis(GO:0061073) |
| 0.0 |
0.1 |
GO:0060332 |
positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 |
0.2 |
GO:0061042 |
vascular wound healing(GO:0061042) |
| 0.0 |
0.0 |
GO:0035669 |
TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 |
0.1 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 |
0.2 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 |
0.1 |
GO:0060681 |
branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 |
0.1 |
GO:0006828 |
manganese ion transport(GO:0006828) |
| 0.0 |
0.5 |
GO:0030947 |
regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 |
0.0 |
GO:0045658 |
regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 |
0.1 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 |
0.1 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 |
0.0 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.0 |
0.1 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.0 |
0.1 |
GO:0036369 |
transcription factor catabolic process(GO:0036369) |
| 0.0 |
0.1 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.1 |
GO:0014809 |
regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 |
0.2 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 |
0.0 |
GO:0038193 |
thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 |
0.1 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.1 |
GO:0032962 |
regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 |
0.1 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.0 |
0.2 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 |
0.1 |
GO:0015705 |
iodide transport(GO:0015705) |
| 0.0 |
0.1 |
GO:0072539 |
T-helper 17 cell differentiation(GO:0072539) |
| 0.0 |
0.1 |
GO:0035026 |
leading edge cell differentiation(GO:0035026) |
| 0.0 |
0.1 |
GO:0006540 |
glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 |
0.2 |
GO:0048003 |
antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 |
0.1 |
GO:0006572 |
tyrosine catabolic process(GO:0006572) |
| 0.0 |
0.1 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 0.0 |
0.2 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.0 |
0.2 |
GO:0035860 |
glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 |
0.1 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 |
0.4 |
GO:0003298 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.6 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.1 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 |
0.0 |
GO:0033591 |
response to L-ascorbic acid(GO:0033591) |
| 0.0 |
0.2 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.2 |
GO:0018298 |
protein-chromophore linkage(GO:0018298) |
| 0.0 |
0.1 |
GO:0019805 |
quinolinate biosynthetic process(GO:0019805) |
| 0.0 |
0.1 |
GO:0072364 |
regulation of cellular ketone metabolic process by regulation of transcription from RNA polymerase II promoter(GO:0072364) |
| 0.0 |
0.1 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 |
0.3 |
GO:0021591 |
ventricular system development(GO:0021591) |
| 0.0 |
0.2 |
GO:0071688 |
striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 |
0.1 |
GO:0002767 |
immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 |
0.1 |
GO:0044245 |
polysaccharide digestion(GO:0044245) |
| 0.0 |
0.3 |
GO:0032098 |
regulation of appetite(GO:0032098) |
| 0.0 |
0.7 |
GO:0005978 |
glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 |
0.7 |
GO:0008088 |
axo-dendritic transport(GO:0008088) |
| 0.0 |
0.5 |
GO:0097178 |
ruffle assembly(GO:0097178) |
| 0.0 |
0.1 |
GO:0035372 |
protein localization to microtubule(GO:0035372) |
| 0.0 |
0.0 |
GO:0010523 |
negative regulation of calcium ion transport into cytosol(GO:0010523) |
| 0.0 |
0.3 |
GO:0001502 |
cartilage condensation(GO:0001502) |
| 0.0 |
0.1 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.0 |
0.2 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.0 |
0.1 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 |
0.0 |
GO:1903597 |
regulation of renal output by angiotensin(GO:0002019) negative regulation of gap junction assembly(GO:1903597) |
| 0.0 |
0.0 |
GO:0003283 |
atrial septum development(GO:0003283) |
| 0.0 |
0.0 |
GO:0097010 |
eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 |
0.4 |
GO:0007050 |
cell cycle arrest(GO:0007050) |
| 0.0 |
0.0 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |