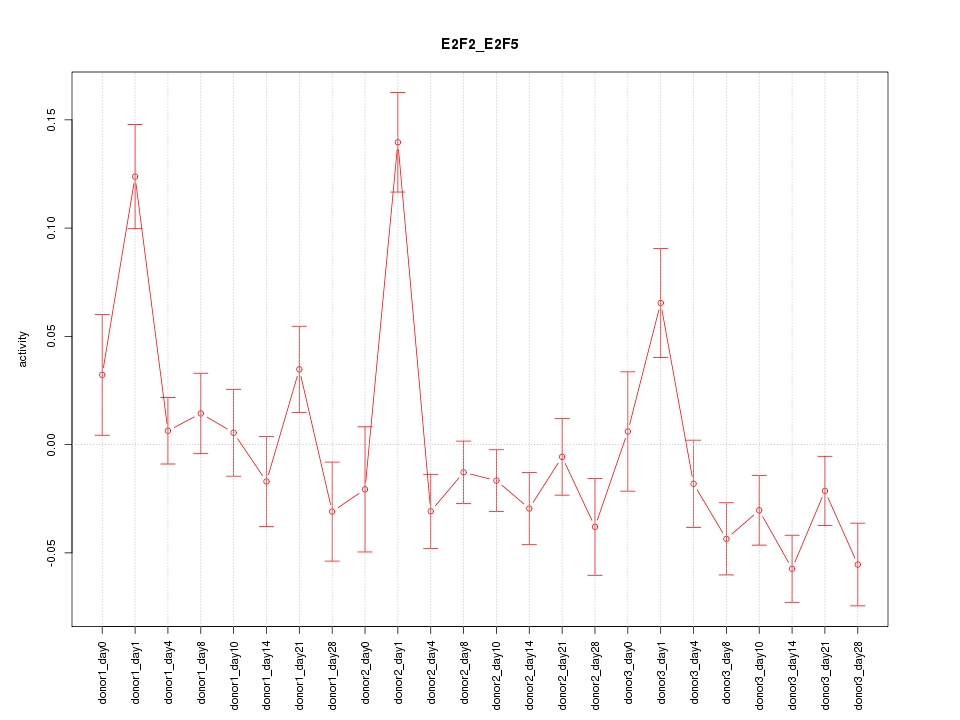
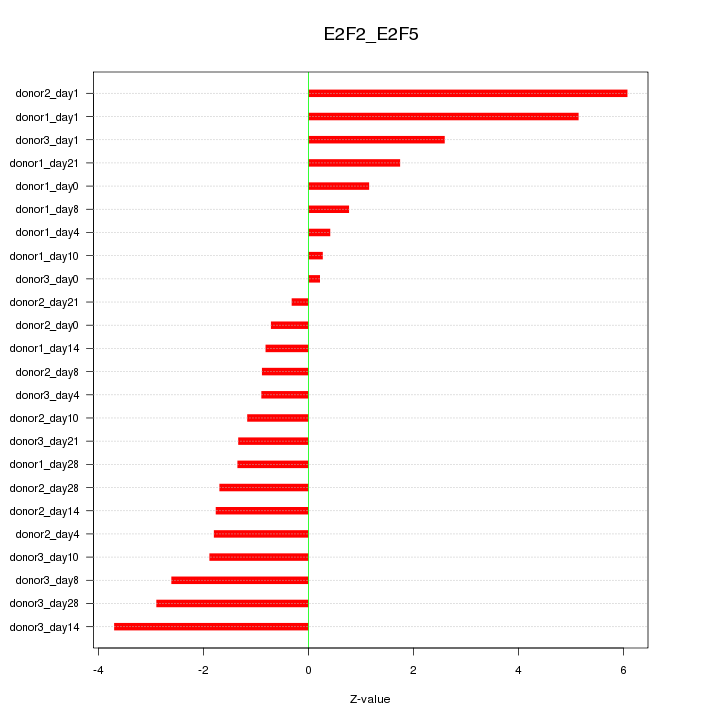
Motif ID: E2F2_E2F5
Z-value: 2.283
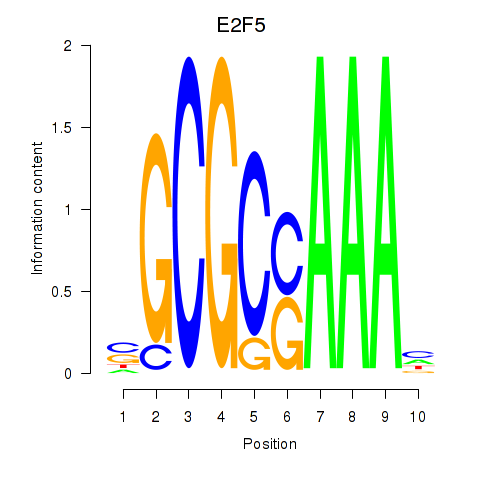
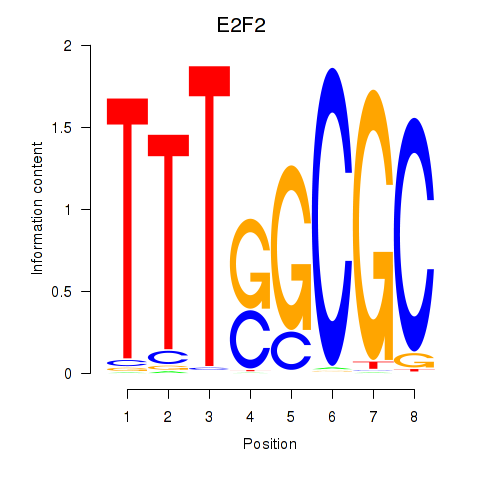
Transcription factors associated with E2F2_E2F5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| E2F2 | ENSG00000007968.6 | E2F2 |
| E2F5 | ENSG00000133740.6 | E2F5 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F2 | hg19_v2_chr1_-_23857698_23857733 | 0.75 | 2.5e-05 | Click! |
| E2F5 | hg19_v2_chr8_+_86121448_86121494 | -0.37 | 7.6e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.6 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 3.4 | 10.2 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 2.9 | 17.3 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 2.9 | 28.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 2.6 | 13.0 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 1.8 | 10.9 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 1.4 | 7.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 1.4 | 4.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 1.3 | 6.7 | GO:0060356 | leucine import(GO:0060356) |
| 1.2 | 6.2 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 1.2 | 4.8 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 1.2 | 7.0 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 1.1 | 4.5 | GO:0006272 | leading strand elongation(GO:0006272) |
| 1.0 | 15.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.8 | 2.4 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.8 | 7.7 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.7 | 2.8 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.7 | 28.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.7 | 2.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.6 | 3.2 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.6 | 6.3 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.6 | 4.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.5 | 2.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.5 | 3.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.5 | 1.4 | GO:0071790 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.5 | 11.8 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.4 | 1.3 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.4 | 1.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.4 | 1.7 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.4 | 1.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.4 | 2.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 2.6 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.3 | 1.8 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.3 | 0.8 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.2 | 2.0 | GO:0033504 | floor plate development(GO:0033504) |
| 0.2 | 1.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.2 | 0.7 | GO:0072186 | metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) |
| 0.2 | 2.2 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.2 | 2.3 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.2 | 0.8 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.2 | 1.0 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.2 | 2.0 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.2 | 1.4 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 | 0.4 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.2 | 1.3 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 | 1.9 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.2 | 0.9 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 1.0 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) |
| 0.2 | 1.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 | 1.9 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 0.8 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.2 | 4.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.2 | 0.6 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.2 | 7.4 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 | 0.3 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 1.6 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 1.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 1.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.5 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 | 5.2 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 1.0 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 0.7 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 5.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 1.2 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 | 11.1 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.1 | 0.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.3 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 1.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 1.3 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.1 | 1.9 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 2.1 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 5.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.3 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.5 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 1.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.2 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 1.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.7 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 1.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.4 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 1.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.9 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.9 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 1.9 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 1.3 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) sensory system development(GO:0048880) |
| 0.0 | 0.5 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.8 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.5 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.0 | 0.5 | GO:1903432 | regulation of TORC1 signaling(GO:1903432) |
| 0.0 | 0.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.9 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 17.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 2.9 | 11.7 | GO:0000811 | GINS complex(GO:0000811) |
| 2.6 | 42.4 | GO:0042555 | MCM complex(GO:0042555) |
| 2.4 | 11.8 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 1.8 | 5.3 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 1.1 | 6.9 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 1.1 | 9.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.9 | 8.5 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.9 | 7.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.8 | 2.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.7 | 7.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.6 | 10.2 | GO:0045120 | pronucleus(GO:0045120) |
| 0.5 | 2.1 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.5 | 1.4 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.5 | 2.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 4.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.4 | 2.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.4 | 6.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.4 | 1.9 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.4 | 1.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 1.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.3 | 3.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.3 | 1.9 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.3 | 5.8 | GO:0000800 | lateral element(GO:0000800) |
| 0.3 | 9.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.3 | 1.0 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.3 | 6.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 0.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) PCNA-p21 complex(GO:0070557) |
| 0.2 | 0.7 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.2 | 1.7 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.5 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.8 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.5 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 6.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.4 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.5 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 1.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 5.9 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.9 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 6.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 5.3 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 13.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.8 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 9.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 4.0 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 2.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 4.6 | GO:0005813 | centrosome(GO:0005813) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 11.8 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.5 | 24.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 1.2 | 7.0 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 1.1 | 9.7 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.8 | 4.8 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.8 | 17.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.7 | 11.7 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.5 | 18.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.5 | 2.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.5 | 7.0 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.5 | 3.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.4 | 6.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.4 | 1.9 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.4 | 1.9 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.4 | 0.7 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.3 | 1.0 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.3 | 2.0 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.3 | 11.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 0.8 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.2 | 1.4 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 1.7 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.2 | 2.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.2 | 1.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.2 | 1.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 1.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 0.6 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 2.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 10.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 7.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 9.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 1.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 2.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 2.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 2.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 18.2 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 2.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 1.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 6.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 6.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 5.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 2.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 13.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 1.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.9 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 3.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 8.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.9 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.8 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 1.3 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 6.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.0 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 4.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 1.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0048038 | quinone binding(GO:0048038) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 27.7 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.7 | 12.9 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 35.4 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.3 | 10.3 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.2 | 5.4 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.2 | 9.9 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.2 | 8.3 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.1 | 2.3 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.1 | 6.6 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 6.4 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.0 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 2.0 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 4.1 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 5.3 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.8 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.0 | 2.0 | PID_PI3K_PLC_TRK_PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.8 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.0 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.8 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.0 | 0.8 | PID_LKB1_PATHWAY | LKB1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 54.1 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 1.6 | 44.0 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 1.0 | 14.5 | REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.6 | 10.6 | REACTOME_ACTIVATION_OF_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.6 | 7.7 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.5 | 11.5 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.4 | 7.4 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.3 | 5.7 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.3 | 1.4 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 11.7 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.2 | 6.7 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 5.3 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.1 | 1.4 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.6 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.6 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.2 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.8 | REACTOME_SPRY_REGULATION_OF_FGF_SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 1.3 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 2.0 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.7 | REACTOME_RNA_POL_I_TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 1.0 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 2.3 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.1 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.9 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.9 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.2 | REACTOME_MRNA_SPLICING_MINOR_PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.8 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.5 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 3.9 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.9 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.9 | REACTOME_M_G1_TRANSITION | Genes involved in M/G1 Transition |
| 0.0 | 0.6 | REACTOME_ABC_FAMILY_PROTEINS_MEDIATED_TRANSPORT | Genes involved in ABC-family proteins mediated transport |