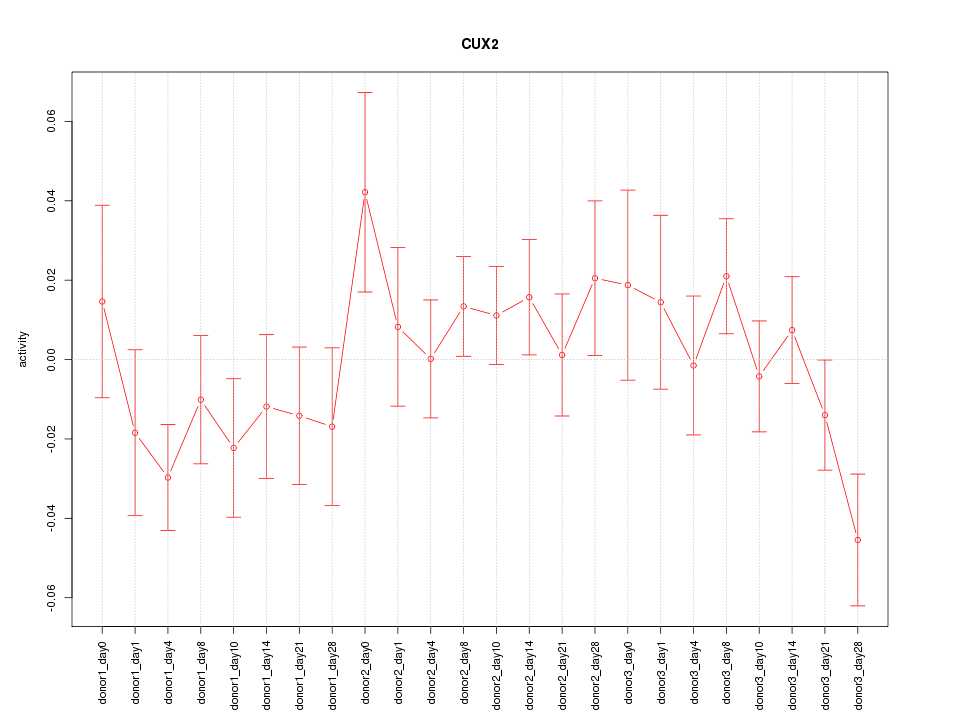
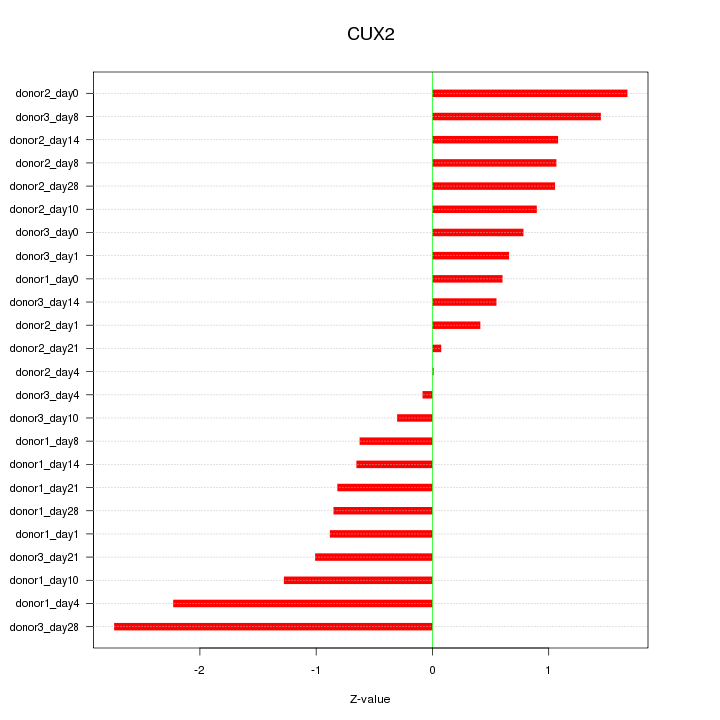
Motif ID: CUX2
Z-value: 1.104
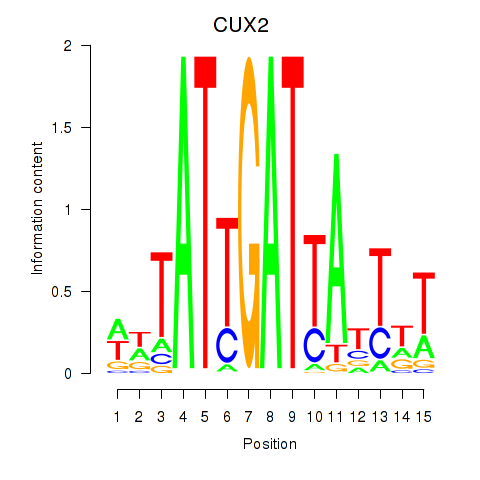
Transcription factors associated with CUX2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| CUX2 | ENSG00000111249.9 | CUX2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CUX2 | hg19_v2_chr12_+_111471828_111471975 | 0.02 | 9.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.6 | 1.7 | GO:0071790 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.4 | 0.4 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.4 | 1.5 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.3 | 1.0 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.3 | 1.0 | GO:2000646 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) positive regulation of receptor catabolic process(GO:2000646) |
| 0.3 | 1.3 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.3 | 1.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.3 | 1.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.3 | 1.3 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.3 | 0.8 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.2 | 0.7 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.2 | 0.6 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.2 | 0.7 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.2 | 0.5 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 1.0 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 1.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.6 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.6 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.7 | GO:1903285 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.7 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.1 | 0.5 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.4 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.1 | 2.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 1.6 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.4 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.8 | GO:2000334 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 1.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 0.4 | GO:0007113 | endomitotic cell cycle(GO:0007113) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 0.7 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 1.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.7 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 0.8 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.3 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.4 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.1 | 0.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.3 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.2 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 1.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.2 | GO:1902725 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 1.1 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.7 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 0.4 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.2 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.6 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.5 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 1.6 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.1 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 1.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.1 | GO:1900005 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.1 | 0.3 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.4 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 | 0.3 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 2.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.3 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.2 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 0.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.3 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.3 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 1.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.4 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 1.0 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 1.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.3 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 1.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 4.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 1.0 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.2 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.5 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.2 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.2 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.0 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.8 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.2 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.1 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.4 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.2 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 1.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.6 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.1 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) regulation of AV node cell action potential(GO:0098904) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 0.4 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 1.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.4 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 2.1 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 1.7 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.4 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.4 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 1.0 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.2 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0032106 | positive regulation of response to extracellular stimulus(GO:0032106) positive regulation of response to nutrient levels(GO:0032109) |
| 0.0 | 0.2 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.4 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.8 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.1 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.5 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.6 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 | 0.5 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.3 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.4 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.8 | GO:0046847 | filopodium assembly(GO:0046847) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.5 | 1.4 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.3 | 1.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.2 | 2.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 1.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 1.0 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 1.0 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.4 | GO:0071749 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.1 | 0.4 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.1 | 1.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.6 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.7 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.6 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 1.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.3 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.7 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 1.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 1.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 4.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 2.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 1.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.6 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 1.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.6 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.7 | GO:0016020 | membrane(GO:0016020) |
| 0.0 | 1.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 1.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.4 | 1.3 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.4 | 1.3 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.4 | 1.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.3 | 1.3 | GO:0030395 | lactose binding(GO:0030395) |
| 0.2 | 1.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.2 | 1.0 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.2 | 1.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 2.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 0.6 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.2 | 1.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 0.5 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 1.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.8 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.4 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 1.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.7 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 0.6 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.5 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 0.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 1.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 1.0 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.3 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 0.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 1.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 1.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 2.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 2.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 1.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.7 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 0.2 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.1 | 1.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.2 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.2 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.1 | 0.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.3 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.2 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 3.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 2.0 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 1.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.9 | PID_S1P_S1P1_PATHWAY | S1P1 pathway |
| 0.0 | 3.2 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.5 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.0 | 1.5 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.0 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.8 | PID_IL27_PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.9 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.0 | 0.6 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.8 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.0 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID_IGF1_PATHWAY | IGF1 pathway |
| 0.0 | 0.6 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.7 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.0 | 0.1 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.1 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.6 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.4 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.7 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.5 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.3 | REACTOME_REVERSIBLE_HYDRATION_OF_CARBON_DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.9 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.1 | REACTOME_GRB2_SOS_PROVIDES_LINKAGE_TO_MAPK_SIGNALING_FOR_INTERGRINS_ | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.1 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.0 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 4.5 | REACTOME_OLFACTORY_SIGNALING_PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 2.2 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_24_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.6 | REACTOME_DOPAMINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.6 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.3 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME_RECYCLING_OF_BILE_ACIDS_AND_SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.0 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.7 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.6 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 2.4 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME_FORMATION_OF_FIBRIN_CLOT_CLOTTING_CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.6 | REACTOME_REGULATION_OF_IFNA_SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.9 | REACTOME_G_ALPHA_Z_SIGNALLING_EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.0 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.3 | REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 2.6 | REACTOME_DIABETES_PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.1 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.2 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.2 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |