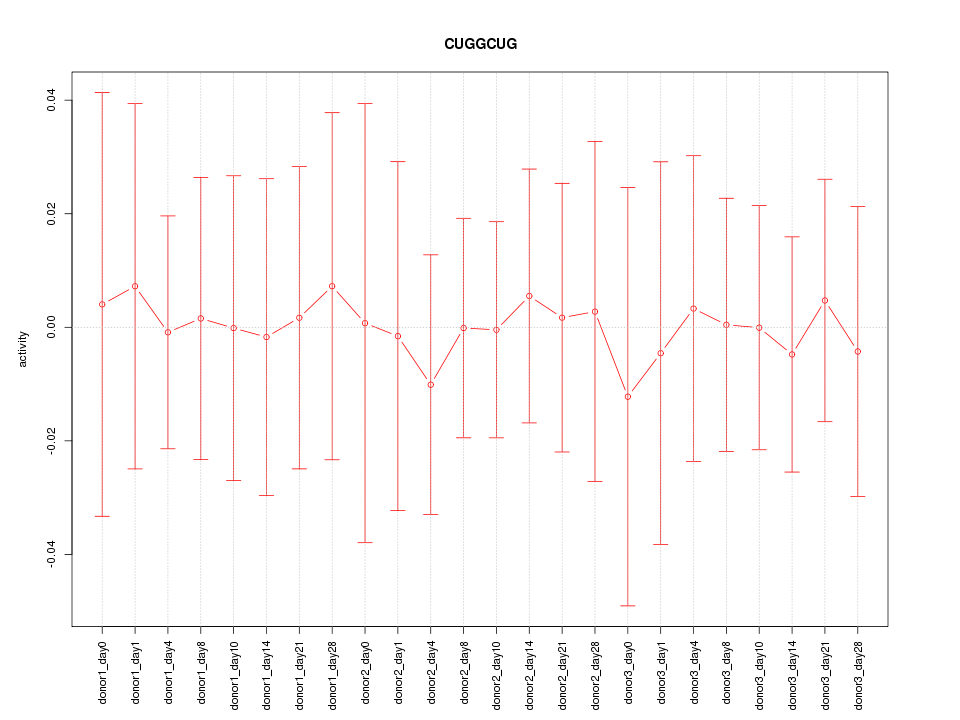
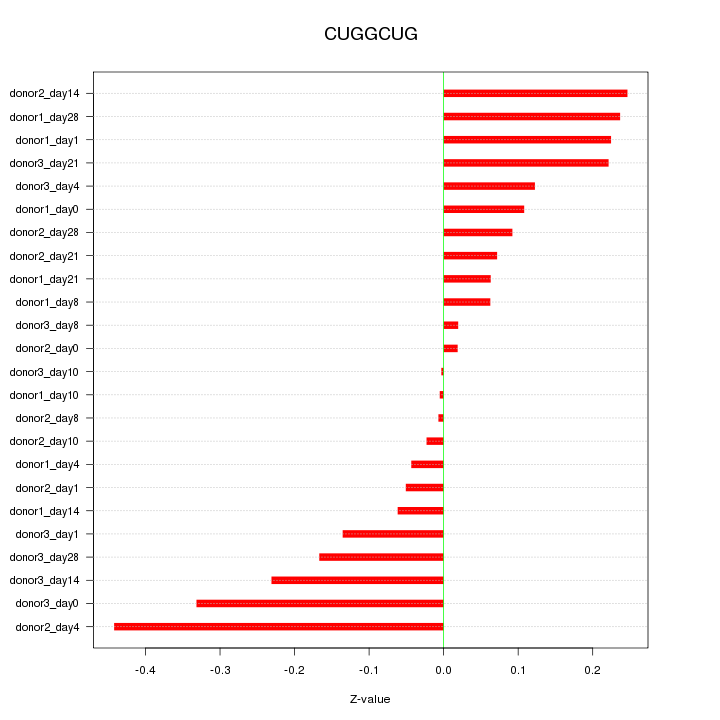
|
chrY_+_15016725
|
0.151
|
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked
|
|
chr2_-_217560248
|
0.128
|
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr11_-_115375107
|
0.108
|
ENST00000545380.1
ENST00000452722.3
ENST00000537058.1
ENST00000536727.1
ENST00000542447.2
ENST00000331581.6
|
CADM1
|
cell adhesion molecule 1
|
|
chr3_-_112360116
|
0.086
|
ENST00000206423.3
ENST00000439685.2
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr14_-_95786200
|
0.084
|
ENST00000298912.4
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr1_+_38022513
|
0.076
|
ENST00000296218.7
|
DNALI1
|
dynein, axonemal, light intermediate chain 1
|
|
chr13_+_24153488
|
0.071
|
ENST00000382258.4
ENST00000382263.3
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr17_+_55333876
|
0.068
|
ENST00000284073.2
|
MSI2
|
musashi RNA-binding protein 2
|
|
chr9_+_129376722
|
0.057
|
ENST00000526117.1
ENST00000373474.4
ENST00000355497.5
ENST00000425646.2
ENST00000561065.1
|
LMX1B
|
LIM homeobox transcription factor 1, beta
|
|
chr16_-_86588627
|
0.056
|
ENST00000565482.1
ENST00000564364.1
ENST00000561989.1
ENST00000543303.2
ENST00000381214.5
ENST00000360900.6
ENST00000322911.6
ENST00000546093.1
ENST00000569000.1
ENST00000562994.1
ENST00000561522.1
|
MTHFSD
|
methenyltetrahydrofolate synthetase domain containing
|
|
chr13_-_61989655
|
0.051
|
ENST00000409204.4
|
PCDH20
|
protocadherin 20
|
|
chr3_-_12200851
|
0.048
|
ENST00000287814.4
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|
|
chr17_-_33700626
|
0.047
|
ENST00000308377.4
ENST00000394566.1
ENST00000589811.1
ENST00000430814.1
ENST00000427966.1
ENST00000588579.1
ENST00000591682.1
ENST00000441608.2
ENST00000592122.1
|
SLFN11
|
schlafen family member 11
|
|
chr1_-_57045228
|
0.045
|
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr11_+_45868957
|
0.044
|
ENST00000443527.2
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr1_-_26633067
|
0.043
|
ENST00000421827.2
ENST00000374215.1
ENST00000374223.1
ENST00000357089.4
ENST00000535108.1
ENST00000314675.7
ENST00000436301.2
ENST00000423664.1
ENST00000374221.3
|
UBXN11
|
UBX domain protein 11
|
|
chrX_+_16964794
|
0.043
|
ENST00000357277.3
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr8_+_75896731
|
0.043
|
ENST00000262207.4
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr8_+_26435359
|
0.042
|
ENST00000311151.5
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chrX_+_9431324
|
0.041
|
ENST00000407597.2
ENST00000424279.1
ENST00000536365.1
ENST00000441088.1
ENST00000380961.1
ENST00000415293.1
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr2_+_131113580
|
0.041
|
ENST00000175756.5
|
PTPN18
|
protein tyrosine phosphatase, non-receptor type 18 (brain-derived)
|
|
chr17_+_29718642
|
0.039
|
ENST00000325874.8
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr12_+_54943134
|
0.038
|
ENST00000243052.3
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent
|
|
chr21_-_43346790
|
0.036
|
ENST00000329623.7
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr12_-_58240470
|
0.036
|
ENST00000548823.1
ENST00000398073.2
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2
|
|
chr5_-_121413974
|
0.035
|
ENST00000231004.4
|
LOX
|
lysyl oxidase
|
|
chr17_-_33775760
|
0.035
|
ENST00000534689.1
ENST00000532210.1
ENST00000526861.1
ENST00000531588.1
ENST00000285013.6
|
SLFN13
|
schlafen family member 13
|
|
chr4_+_77356248
|
0.035
|
ENST00000296043.6
|
SHROOM3
|
shroom family member 3
|
|
chr1_+_11333245
|
0.033
|
ENST00000376810.5
|
UBIAD1
|
UbiA prenyltransferase domain containing 1
|
|
chr11_-_72504637
|
0.033
|
ENST00000359373.5
ENST00000536377.1
|
ARAP1
STARD10
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
StAR-related lipid transfer (START) domain containing 10
|
|
chr16_-_17564738
|
0.032
|
ENST00000261381.6
|
XYLT1
|
xylosyltransferase I
|
|
chr5_-_132948216
|
0.030
|
ENST00000265342.7
|
FSTL4
|
follistatin-like 4
|
|
chr19_-_14316980
|
0.029
|
ENST00000361434.3
ENST00000340736.6
|
LPHN1
|
latrophilin 1
|
|
chr19_+_41816053
|
0.029
|
ENST00000269967.3
|
CCDC97
|
coiled-coil domain containing 97
|
|
chr12_-_50297638
|
0.028
|
ENST00000320634.3
|
FAIM2
|
Fas apoptotic inhibitory molecule 2
|
|
chr19_+_13135386
|
0.028
|
ENST00000360105.4
ENST00000588228.1
ENST00000591028.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr12_-_54673871
|
0.028
|
ENST00000209875.4
|
CBX5
|
chromobox homolog 5
|
|
chr6_-_44281043
|
0.027
|
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial
|
|
chr11_+_63706444
|
0.027
|
ENST00000377793.4
ENST00000456907.2
ENST00000539656.1
|
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit
|
|
chr13_+_27131887
|
0.027
|
ENST00000335327.5
|
WASF3
|
WAS protein family, member 3
|
|
chr5_+_133861790
|
0.026
|
ENST00000395003.1
|
PHF15
|
jade family PHD finger 2
|
|
chr5_-_146833485
|
0.026
|
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr1_+_36348790
|
0.026
|
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1
|
|
chr1_+_150122034
|
0.025
|
ENST00000025469.6
ENST00000369124.4
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr1_-_109940550
|
0.023
|
ENST00000256637.6
|
SORT1
|
sortilin 1
|
|
chr1_-_1677358
|
0.023
|
ENST00000355439.2
ENST00000400924.1
ENST00000246421.4
|
SLC35E2
|
solute carrier family 35, member E2
|
|
chr17_-_27893990
|
0.023
|
ENST00000307201.4
|
ABHD15
|
abhydrolase domain containing 15
|
|
chr17_-_1928621
|
0.022
|
ENST00000331238.6
|
RTN4RL1
|
reticulon 4 receptor-like 1
|
|
chr1_+_210501589
|
0.022
|
ENST00000413764.2
ENST00000541565.1
|
HHAT
|
hedgehog acyltransferase
|
|
chr12_-_57400227
|
0.022
|
ENST00000300101.2
|
ZBTB39
|
zinc finger and BTB domain containing 39
|
|
chr9_-_111882195
|
0.021
|
ENST00000374586.3
|
TMEM245
|
transmembrane protein 245
|
|
chr1_+_15943995
|
0.021
|
ENST00000480945.1
|
DDI2
|
DNA-damage inducible 1 homolog 2 (S. cerevisiae)
|
|
chr4_+_72204755
|
0.021
|
ENST00000512686.1
ENST00000340595.3
|
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4
|
|
chr2_+_149632783
|
0.020
|
ENST00000435030.1
|
KIF5C
|
kinesin family member 5C
|
|
chr20_+_32581452
|
0.019
|
ENST00000375114.3
ENST00000448364.1
|
RALY
|
RALY heterogeneous nuclear ribonucleoprotein
|
|
chr1_+_153700518
|
0.019
|
ENST00000318967.2
ENST00000456435.1
ENST00000435409.2
|
INTS3
|
integrator complex subunit 3
|
|
chr5_+_153825510
|
0.019
|
ENST00000297109.6
|
SAP30L
|
SAP30-like
|
|
chr9_+_95087766
|
0.019
|
ENST00000375587.3
|
CENPP
|
centromere protein P
|
|
chr11_-_73309228
|
0.018
|
ENST00000356467.4
ENST00000064778.4
|
FAM168A
|
family with sequence similarity 168, member A
|
|
chr20_-_41818373
|
0.018
|
ENST00000373187.1
ENST00000356100.2
ENST00000373184.1
ENST00000373190.1
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chr1_+_27561007
|
0.018
|
ENST00000319394.3
|
WDTC1
|
WD and tetratricopeptide repeats 1
|
|
chr17_+_57408994
|
0.018
|
ENST00000312655.4
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chr22_+_39795746
|
0.018
|
ENST00000216160.6
ENST00000331454.3
|
TAB1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1
|
|
chr5_-_133512683
|
0.018
|
ENST00000353411.6
|
SKP1
|
S-phase kinase-associated protein 1
|
|
chr12_-_105630016
|
0.018
|
ENST00000258530.3
|
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2
|
|
chr12_-_82153087
|
0.017
|
ENST00000547623.1
ENST00000549396.1
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr1_+_206557366
|
0.017
|
ENST00000414007.1
ENST00000419187.2
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2
|
|
chr7_+_32535060
|
0.017
|
ENST00000318709.4
ENST00000409301.1
ENST00000404479.1
|
AVL9
|
AVL9 homolog (S. cerevisiase)
|
|
chr19_-_14201507
|
0.017
|
ENST00000533683.2
|
SAMD1
|
sterile alpha motif domain containing 1
|
|
chr19_-_2721412
|
0.017
|
ENST00000323469.4
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr12_-_59313270
|
0.017
|
ENST00000379141.4
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr8_-_119124045
|
0.016
|
ENST00000378204.2
|
EXT1
|
exostosin glycosyltransferase 1
|
|
chr22_-_39239987
|
0.016
|
ENST00000333039.2
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr1_+_32645269
|
0.016
|
ENST00000373610.3
|
TXLNA
|
taxilin alpha
|
|
chr3_-_133614597
|
0.016
|
ENST00000285208.4
ENST00000460865.3
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr11_-_106889250
|
0.015
|
ENST00000526355.2
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2
|
|
chr15_+_75498355
|
0.015
|
ENST00000567617.1
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr11_-_69519410
|
0.015
|
ENST00000294312.3
|
FGF19
|
fibroblast growth factor 19
|
|
chr12_-_49449107
|
0.015
|
ENST00000301067.7
|
KMT2D
|
lysine (K)-specific methyltransferase 2D
|
|
chr11_+_123396528
|
0.015
|
ENST00000322282.7
ENST00000529750.1
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr7_-_99036270
|
0.015
|
ENST00000430029.1
ENST00000419981.1
ENST00000292478.4
|
PTCD1
|
pentatricopeptide repeat domain 1
|
|
chr17_+_60536002
|
0.015
|
ENST00000582809.1
|
TLK2
|
tousled-like kinase 2
|
|
chrX_+_70752917
|
0.014
|
ENST00000373719.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase
|
|
chr5_+_6448736
|
0.014
|
ENST00000399816.3
|
UBE2QL1
|
ubiquitin-conjugating enzyme E2Q family-like 1
|
|
chr12_-_56652111
|
0.013
|
ENST00000267116.7
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr12_-_56727487
|
0.013
|
ENST00000548043.1
ENST00000425394.2
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr8_+_21946681
|
0.013
|
ENST00000289921.7
|
FAM160B2
|
family with sequence similarity 160, member B2
|
|
chr14_-_39901618
|
0.013
|
ENST00000554932.1
ENST00000298097.7
|
FBXO33
|
F-box protein 33
|
|
chr2_-_97405775
|
0.013
|
ENST00000264963.4
ENST00000537039.1
ENST00000377079.4
ENST00000426463.2
ENST00000534882.1
|
LMAN2L
|
lectin, mannose-binding 2-like
|
|
chr2_-_162931052
|
0.013
|
ENST00000360534.3
|
DPP4
|
dipeptidyl-peptidase 4
|
|
chr15_-_61521495
|
0.013
|
ENST00000335670.6
|
RORA
|
RAR-related orphan receptor A
|
|
chr13_+_51796497
|
0.013
|
ENST00000322475.8
ENST00000280057.6
|
FAM124A
|
family with sequence similarity 124A
|
|
chr1_-_153931052
|
0.013
|
ENST00000368630.3
ENST00000368633.1
|
CRTC2
|
CREB regulated transcription coactivator 2
|
|
chr14_+_23067146
|
0.012
|
ENST00000428304.2
|
ABHD4
|
abhydrolase domain containing 4
|
|
chr2_+_105471969
|
0.012
|
ENST00000361360.2
|
POU3F3
|
POU class 3 homeobox 3
|
|
chr14_-_75179774
|
0.012
|
ENST00000555249.1
ENST00000556202.1
ENST00000356357.4
ENST00000338772.5
|
AREL1
AC007956.1
|
apoptosis resistant E3 ubiquitin protein ligase 1
Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein
|
|
chr16_-_65155833
|
0.012
|
ENST00000566827.1
ENST00000394156.3
ENST00000562998.1
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr12_-_14133053
|
0.012
|
ENST00000609686.1
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr10_-_32636106
|
0.012
|
ENST00000263062.8
ENST00000319778.6
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr10_-_81965307
|
0.012
|
ENST00000537102.1
ENST00000372231.3
ENST00000438331.1
ENST00000422982.3
ENST00000360615.4
ENST00000265447.4
|
ANXA11
|
annexin A11
|
|
chr2_+_131513864
|
0.012
|
ENST00000458606.1
ENST00000423981.1
|
AMER3
|
APC membrane recruitment protein 3
|
|
chr7_+_77325738
|
0.012
|
ENST00000334955.8
|
RSBN1L
|
round spermatid basic protein 1-like
|
|
chr14_+_57735614
|
0.011
|
ENST00000261558.3
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit
|
|
chr15_-_65579177
|
0.011
|
ENST00000444347.2
ENST00000261888.6
|
PARP16
|
poly (ADP-ribose) polymerase family, member 16
|
|
chr12_-_58027138
|
0.011
|
ENST00000341156.4
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1
|
|
chr7_+_66386204
|
0.011
|
ENST00000341567.4
ENST00000607045.1
|
TMEM248
|
transmembrane protein 248
|
|
chr11_+_71640071
|
0.011
|
ENST00000533380.1
ENST00000393713.3
ENST00000545854.1
|
RNF121
|
ring finger protein 121
|
|
chr2_+_134877740
|
0.010
|
ENST00000409645.1
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase
|
|
chr22_-_19435755
|
0.010
|
ENST00000542103.1
ENST00000399562.4
|
C22orf39
|
chromosome 22 open reading frame 39
|
|
chr16_-_71758602
|
0.010
|
ENST00000568954.1
|
PHLPP2
|
PH domain and leucine rich repeat protein phosphatase 2
|
|
chr6_+_42981922
|
0.010
|
ENST00000326974.4
ENST00000244670.8
|
KLHDC3
|
kelch domain containing 3
|
|
chr15_-_86338134
|
0.010
|
ENST00000337975.5
|
KLHL25
|
kelch-like family member 25
|
|
chr17_+_16318850
|
0.010
|
ENST00000338560.7
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr17_+_61699766
|
0.010
|
ENST00000579585.1
ENST00000584573.1
ENST00000361733.3
ENST00000361357.3
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr20_+_34824355
|
0.009
|
ENST00000397286.3
ENST00000320849.4
ENST00000373932.3
|
AAR2
|
AAR2 splicing factor homolog (S. cerevisiae)
|
|
chr2_+_44066101
|
0.009
|
ENST00000272286.2
|
ABCG8
|
ATP-binding cassette, sub-family G (WHITE), member 8
|
|
chr18_-_45456930
|
0.009
|
ENST00000262160.6
ENST00000587269.1
|
SMAD2
|
SMAD family member 2
|
|
chr2_+_68384976
|
0.009
|
ENST00000263657.2
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae)
|
|
chr19_+_3359561
|
0.009
|
ENST00000589123.1
ENST00000346156.5
ENST00000395111.3
ENST00000586919.1
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr10_+_73724123
|
0.009
|
ENST00000373115.4
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3
|
|
chr3_+_187930719
|
0.009
|
ENST00000312675.4
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr12_-_498620
|
0.009
|
ENST00000399788.2
ENST00000382815.4
|
KDM5A
|
lysine (K)-specific demethylase 5A
|
|
chr2_-_239197201
|
0.009
|
ENST00000254658.3
|
PER2
|
period circadian clock 2
|
|
chr11_+_82868030
|
0.008
|
ENST00000298281.4
ENST00000530660.1
|
PCF11
|
PCF11 cleavage and polyadenylation factor subunit
|
|
chr11_-_62572901
|
0.008
|
ENST00000439713.2
ENST00000531131.1
ENST00000530875.1
ENST00000531709.2
ENST00000294172.2
|
NXF1
|
nuclear RNA export factor 1
|
|
chr17_+_35294075
|
0.008
|
ENST00000254457.5
|
LHX1
|
LIM homeobox 1
|
|
chr3_-_58196688
|
0.008
|
ENST00000486455.1
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr1_-_48462566
|
0.008
|
ENST00000606738.2
|
TRABD2B
|
TraB domain containing 2B
|
|
chr11_+_120382417
|
0.008
|
ENST00000527524.2
ENST00000375081.2
|
GRIK4
AP002348.1
|
glutamate receptor, ionotropic, kainate 4
Uncharacterized protein
|
|
chr1_+_112162381
|
0.008
|
ENST00000433097.1
ENST00000369709.3
ENST00000436150.2
|
RAP1A
|
RAP1A, member of RAS oncogene family
|
|
chr17_-_27916621
|
0.008
|
ENST00000225394.3
|
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1
|
|
chr11_-_62521614
|
0.008
|
ENST00000527994.1
ENST00000394807.3
|
ZBTB3
|
zinc finger and BTB domain containing 3
|
|
chr1_-_155211017
|
0.008
|
ENST00000536770.1
ENST00000368373.3
|
GBA
|
glucosidase, beta, acid
|
|
chr8_-_75233563
|
0.008
|
ENST00000342232.4
|
JPH1
|
junctophilin 1
|
|
chr21_+_42539701
|
0.008
|
ENST00000330333.6
ENST00000328735.6
ENST00000347667.5
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr2_-_68479614
|
0.008
|
ENST00000234310.3
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha
|
|
chr10_-_61666267
|
0.007
|
ENST00000263102.6
|
CCDC6
|
coiled-coil domain containing 6
|
|
chr6_-_166075557
|
0.007
|
ENST00000539869.2
ENST00000366882.1
|
PDE10A
|
phosphodiesterase 10A
|
|
chr1_+_201798269
|
0.007
|
ENST00000361565.4
|
IPO9
|
importin 9
|
|
chr6_+_36853607
|
0.007
|
ENST00000480824.2
ENST00000355190.3
ENST00000373685.1
|
C6orf89
|
chromosome 6 open reading frame 89
|
|
chr20_-_35374456
|
0.007
|
ENST00000373803.2
ENST00000359675.2
ENST00000540765.1
ENST00000349004.1
|
NDRG3
|
NDRG family member 3
|
|
chr19_+_57019212
|
0.007
|
ENST00000308031.5
ENST00000591537.1
|
ZNF471
|
zinc finger protein 471
|
|
chr12_+_52306113
|
0.007
|
ENST00000547400.1
ENST00000550683.1
ENST00000419526.2
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr19_+_34745442
|
0.007
|
ENST00000299505.6
ENST00000588470.1
ENST00000589583.1
ENST00000588338.2
|
KIAA0355
|
KIAA0355
|
|
chr7_+_30323923
|
0.006
|
ENST00000323037.4
|
ZNRF2
|
zinc and ring finger 2
|
|
chr16_+_31085714
|
0.006
|
ENST00000300850.5
ENST00000564189.1
ENST00000428260.1
|
ZNF646
|
zinc finger protein 646
|
|
chr22_+_20067738
|
0.006
|
ENST00000351989.3
ENST00000383024.2
|
DGCR8
|
DGCR8 microprocessor complex subunit
|
|
chr8_-_21646311
|
0.006
|
ENST00000524240.1
ENST00000400782.4
|
GFRA2
|
GDNF family receptor alpha 2
|
|
chr1_-_26372602
|
0.006
|
ENST00000374278.3
ENST00000374276.3
|
SLC30A2
|
solute carrier family 30 (zinc transporter), member 2
|
|
chr20_-_48184638
|
0.006
|
ENST00000244043.4
|
PTGIS
|
prostaglandin I2 (prostacyclin) synthase
|
|
chr17_-_60142609
|
0.006
|
ENST00000397786.2
|
MED13
|
mediator complex subunit 13
|
|
chr2_+_204192942
|
0.006
|
ENST00000295851.5
ENST00000261017.5
|
ABI2
|
abl-interactor 2
|
|
chr22_-_42828375
|
0.006
|
ENST00000329021.5
|
NFAM1
|
NFAT activating protein with ITAM motif 1
|
|
chr17_+_47653178
|
0.006
|
ENST00000328741.5
|
NXPH3
|
neurexophilin 3
|
|
chr16_-_66730583
|
0.006
|
ENST00000330687.4
ENST00000394106.2
ENST00000563952.1
|
CMTM4
|
CKLF-like MARVEL transmembrane domain containing 4
|
|
chr16_+_68298405
|
0.006
|
ENST00000219343.6
ENST00000566834.1
ENST00000566454.1
|
SLC7A6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6
|
|
chr20_+_3451650
|
0.006
|
ENST00000262919.5
|
ATRN
|
attractin
|
|
chr20_+_33292068
|
0.005
|
ENST00000374810.3
ENST00000374809.2
ENST00000451665.1
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr10_+_104678032
|
0.005
|
ENST00000369878.4
ENST00000369875.3
|
CNNM2
|
cyclin M2
|
|
chr1_-_114355083
|
0.005
|
ENST00000261441.5
|
RSBN1
|
round spermatid basic protein 1
|
|
chr2_+_25015968
|
0.005
|
ENST00000380834.2
ENST00000473706.1
|
CENPO
|
centromere protein O
|
|
chrX_+_154254960
|
0.005
|
ENST00000369498.3
|
FUNDC2
|
FUN14 domain containing 2
|
|
chr10_-_70287231
|
0.005
|
ENST00000609923.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16
|
|
chr10_+_102295616
|
0.005
|
ENST00000299163.6
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor
|
|
chr1_-_37499726
|
0.004
|
ENST00000373091.3
ENST00000373093.4
|
GRIK3
|
glutamate receptor, ionotropic, kainate 3
|
|
chr7_-_150864635
|
0.004
|
ENST00000297537.4
|
GBX1
|
gastrulation brain homeobox 1
|
|
chr1_+_16693578
|
0.004
|
ENST00000401088.4
ENST00000471507.1
ENST00000401089.3
ENST00000375590.3
ENST00000492354.1
|
SZRD1
|
SUZ RNA binding domain containing 1
|
|
chr12_-_9102549
|
0.004
|
ENST00000000412.3
|
M6PR
|
mannose-6-phosphate receptor (cation dependent)
|
|
chr1_-_17766198
|
0.004
|
ENST00000375436.4
|
RCC2
|
regulator of chromosome condensation 2
|
|
chr8_+_145743360
|
0.004
|
ENST00000527730.1
ENST00000529022.1
ENST00000292524.1
|
LRRC14
|
leucine rich repeat containing 14
|
|
chr6_-_44265411
|
0.004
|
ENST00000371505.4
|
TCTE1
|
t-complex-associated-testis-expressed 1
|
|
chr20_+_48552908
|
0.004
|
ENST00000244061.2
|
RNF114
|
ring finger protein 114
|
|
chr10_-_25012597
|
0.004
|
ENST00000396432.2
|
ARHGAP21
|
Rho GTPase activating protein 21
|
|
chr1_+_33546714
|
0.004
|
ENST00000294517.6
ENST00000358680.3
ENST00000373443.3
ENST00000398167.1
|
ADC
|
arginine decarboxylase
|
|
chr19_-_55881741
|
0.003
|
ENST00000264563.2
ENST00000590625.1
ENST00000585513.1
|
IL11
|
interleukin 11
|
|
chr14_+_63671105
|
0.003
|
ENST00000316754.3
|
RHOJ
|
ras homolog family member J
|
|
chr5_+_139175380
|
0.003
|
ENST00000274710.3
|
PSD2
|
pleckstrin and Sec7 domain containing 2
|
|
chr2_-_69870835
|
0.003
|
ENST00000409085.4
ENST00000406297.3
|
AAK1
|
AP2 associated kinase 1
|
|
chr3_-_47555167
|
0.003
|
ENST00000296149.4
|
ELP6
|
elongator acetyltransferase complex subunit 6
|
|
chr1_+_159141397
|
0.003
|
ENST00000368124.4
ENST00000368125.4
ENST00000416746.1
|
CADM3
|
cell adhesion molecule 3
|
|
chr5_+_60628074
|
0.003
|
ENST00000252744.5
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr7_-_5463175
|
0.003
|
ENST00000399537.4
ENST00000430969.1
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr10_-_88281494
|
0.003
|
ENST00000298767.5
|
WAPAL
|
wings apart-like homolog (Drosophila)
|
|
chr16_-_71843047
|
0.003
|
ENST00000299980.4
ENST00000393512.3
|
AP1G1
|
adaptor-related protein complex 1, gamma 1 subunit
|
|
chrX_+_119495934
|
0.003
|
ENST00000218008.3
ENST00000361319.3
ENST00000539306.1
|
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide
|
|
chr14_+_23845995
|
0.003
|
ENST00000359320.3
|
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5
|
|
chr17_+_65821780
|
0.003
|
ENST00000321892.4
ENST00000335221.5
ENST00000306378.6
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr16_+_16043406
|
0.003
|
ENST00000399410.3
ENST00000399408.2
ENST00000346370.5
ENST00000351154.5
ENST00000345148.5
ENST00000349029.5
|
ABCC1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 1
|
|
chr11_+_119205222
|
0.003
|
ENST00000311413.4
|
RNF26
|
ring finger protein 26
|
|
chr5_-_160973649
|
0.003
|
ENST00000393959.1
ENST00000517547.1
|
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2
|
|
chr14_-_47812321
|
0.003
|
ENST00000357362.3
ENST00000486952.2
ENST00000426342.1
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr13_+_28813645
|
0.003
|
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr1_-_33430286
|
0.003
|
ENST00000373456.7
ENST00000356990.5
ENST00000235150.4
|
RNF19B
|
ring finger protein 19B
|
|
chr22_+_26138108
|
0.002
|
ENST00000536101.1
ENST00000335473.7
ENST00000407587.2
|
MYO18B
|
myosin XVIIIB
|
|
chrX_-_152864632
|
0.002
|
ENST00000406277.2
|
FAM58A
|
family with sequence similarity 58, member A
|
|
chr1_-_35325400
|
0.002
|
ENST00000521580.2
|
SMIM12
|
small integral membrane protein 12
|
|
chr19_-_913160
|
0.002
|
ENST00000361574.5
ENST00000587975.1
|
R3HDM4
|
R3H domain containing 4
|
|
chr2_+_73114489
|
0.002
|
ENST00000234454.5
|
SPR
|
sepiapterin reductase (7,8-dihydrobiopterin:NADP+ oxidoreductase)
|
|
chr11_+_66610883
|
0.002
|
ENST00000309657.3
ENST00000524506.1
|
RCE1
|
Ras converting CAAX endopeptidase 1
|
|
chr19_-_14228541
|
0.002
|
ENST00000590853.1
ENST00000308677.4
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|