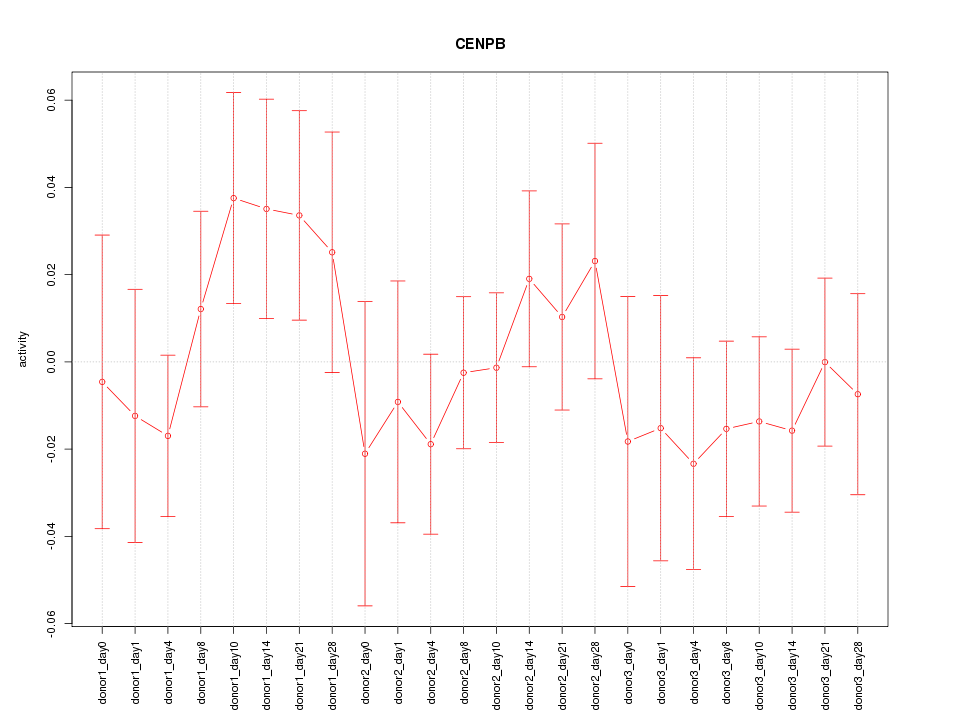
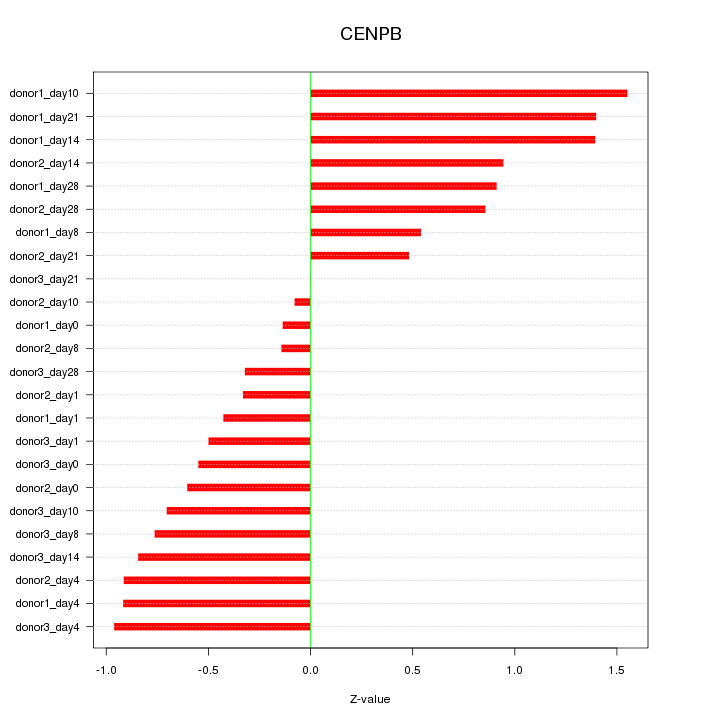
Motif ID: CENPB
Z-value: 0.792
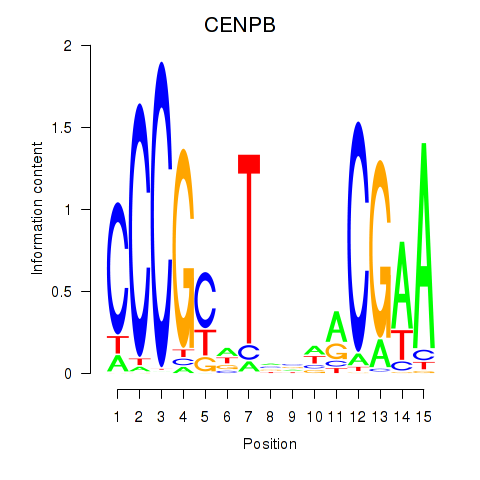
Transcription factors associated with CENPB:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| CENPB | ENSG00000125817.7 | CENPB |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CENPB | hg19_v2_chr20_-_3767324_3767443 | 0.05 | 8.3e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.1 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.5 | 2.2 | GO:0072134 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme morphogenesis(GO:0072134) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) allantois development(GO:1905069) |
| 0.3 | 0.9 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.2 | 0.5 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.2 | 0.8 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.2 | 1.0 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.2 | 0.5 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.8 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.3 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.2 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.1 | 0.4 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 1.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 1.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.2 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.0 | 0.9 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 5.3 | GO:0001942 | hair follicle development(GO:0001942) |
| 0.0 | 6.0 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.6 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.2 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.2 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.4 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.5 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.4 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 2.3 | GO:0014902 | myotube differentiation(GO:0014902) |
| 0.0 | 0.8 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 6.0 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 1.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.6 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.9 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 11.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.4 | 1.3 | GO:0004324 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.2 | 6.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 2.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 0.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.4 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.4 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.8 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.5 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 6.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 1.0 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.1 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.5 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 1.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.7 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 1.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.0 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.2 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.1 | 2.3 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.9 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.9 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.0 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.9 | REACTOME_REGULATED_PROTEOLYSIS_OF_P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.3 | REACTOME_TETRAHYDROBIOPTERIN_BH4_SYNTHESIS_RECYCLING_SALVAGE_AND_REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.9 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.5 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.0 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |