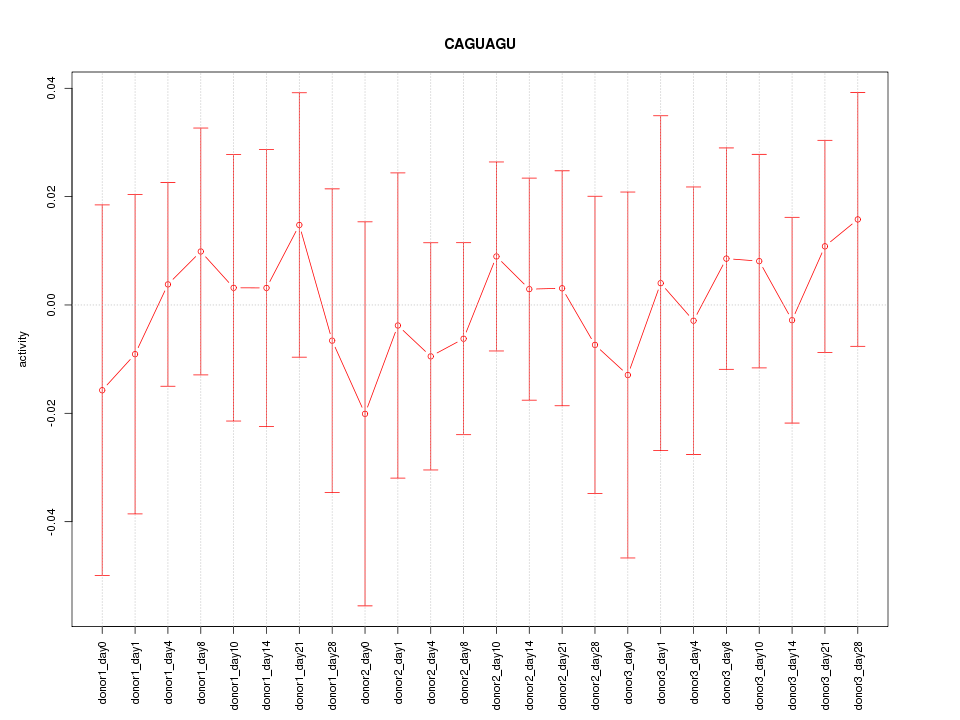
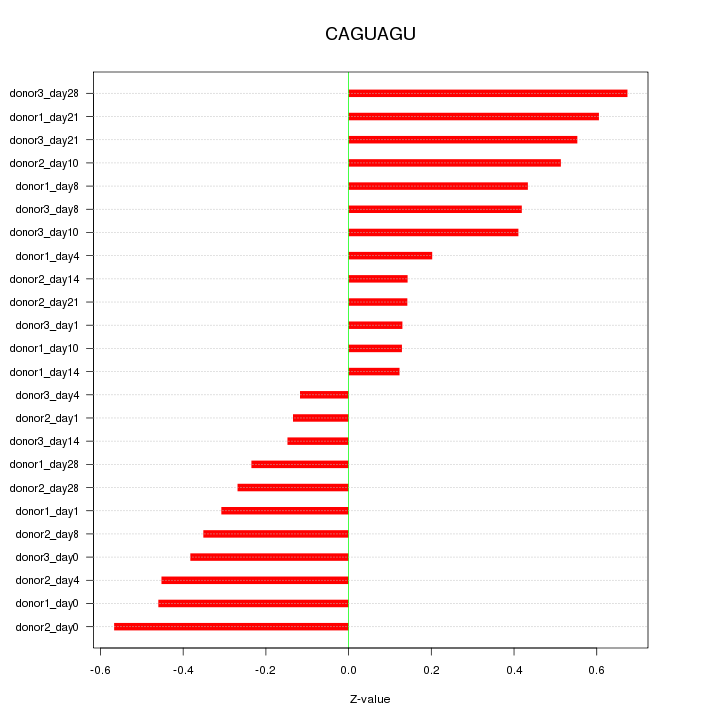
| 0.1 |
0.3 |
GO:0002071 |
glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.1 |
0.3 |
GO:0015993 |
L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 |
0.3 |
GO:0003099 |
vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.1 |
0.2 |
GO:2000364 |
regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.1 |
0.2 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.1 |
0.4 |
GO:1901341 |
activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 |
0.2 |
GO:0044205 |
'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 |
0.3 |
GO:2000568 |
memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 |
0.2 |
GO:0006391 |
transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 |
0.3 |
GO:0060005 |
vestibular reflex(GO:0060005) |
| 0.0 |
0.2 |
GO:1904046 |
negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 |
0.1 |
GO:0035669 |
TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 |
0.3 |
GO:1903385 |
regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 |
0.2 |
GO:0061428 |
negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 |
0.5 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.0 |
0.1 |
GO:1900155 |
regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 |
0.5 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.2 |
GO:1903826 |
arginine transmembrane transport(GO:1903826) |
| 0.0 |
0.1 |
GO:0007185 |
transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 |
0.5 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.0 |
0.2 |
GO:0043129 |
surfactant homeostasis(GO:0043129) |
| 0.0 |
0.1 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.0 |
0.2 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.0 |
0.2 |
GO:0071896 |
protein localization to adherens junction(GO:0071896) |
| 0.0 |
0.1 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 |
0.2 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 |
0.1 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 |
0.1 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.0 |
0.1 |
GO:0042713 |
sperm ejaculation(GO:0042713) |
| 0.0 |
0.2 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.0 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 |
0.1 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 |
0.1 |
GO:0016554 |
cytidine to uridine editing(GO:0016554) |
| 0.0 |
0.0 |
GO:1900169 |
regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 |
0.1 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.0 |
0.1 |
GO:0045875 |
negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 |
0.4 |
GO:0072189 |
ureter development(GO:0072189) |
| 0.0 |
0.0 |
GO:1903371 |
regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 |
0.1 |
GO:0061687 |
detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 |
0.1 |
GO:0019075 |
virus maturation(GO:0019075) |
| 0.0 |
0.0 |
GO:0033140 |
negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 |
0.1 |
GO:2000546 |
positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 |
0.2 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.0 |
0.1 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 |
0.1 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |