Motif ID: BARHL1
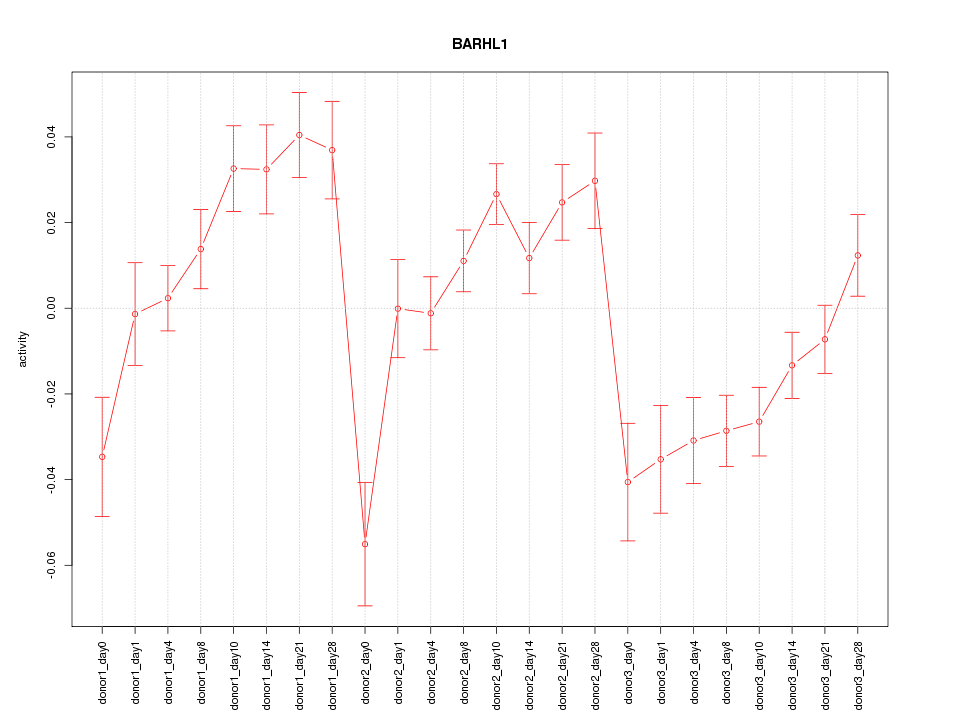
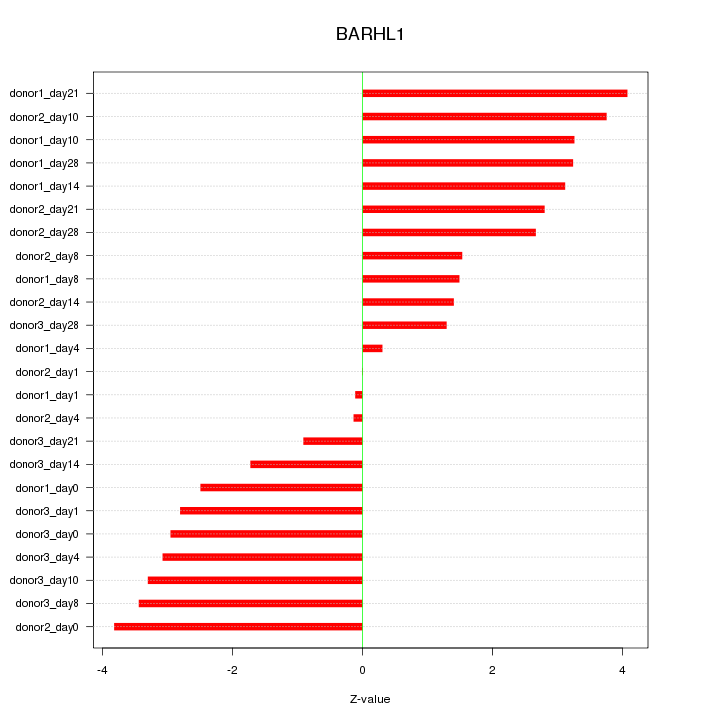
Z-value: 2.570
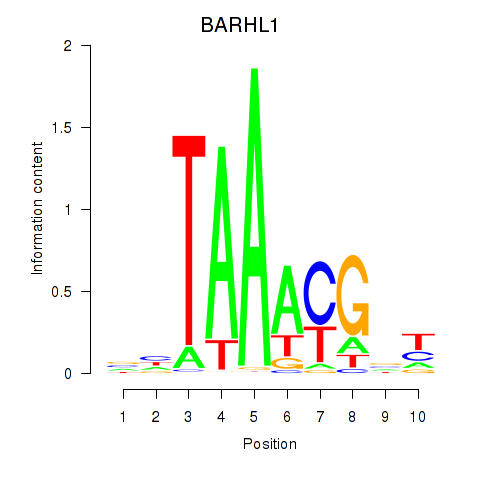
Transcription factors associated with BARHL1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| BARHL1 | ENSG00000125492.5 | BARHL1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BARHL1 | hg19_v2_chr9_+_135457530_135457572 | -0.52 | 8.8e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 8.9 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 2.5 | 10.0 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 2.4 | 17.0 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 2.1 | 6.3 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 1.8 | 1.8 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 1.6 | 1.6 | GO:0045176 | apical protein localization(GO:0045176) |
| 1.6 | 15.6 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 1.4 | 5.7 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 1.3 | 5.3 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 1.3 | 5.2 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 1.2 | 4.9 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 1.2 | 4.7 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 1.1 | 12.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 1.1 | 4.5 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 1.1 | 7.9 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 1.1 | 2.2 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 1.1 | 23.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 1.1 | 4.3 | GO:0042710 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 1.0 | 3.1 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 1.0 | 3.1 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 1.0 | 2.9 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.9 | 5.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.9 | 3.7 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.9 | 17.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.9 | 0.9 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.8 | 0.8 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.8 | 6.7 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.8 | 0.8 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.8 | 3.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.8 | 2.3 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.7 | 0.7 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.7 | 2.9 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.7 | 3.6 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.7 | 2.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.7 | 6.4 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.7 | 2.8 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.7 | 13.9 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.7 | 2.1 | GO:2001301 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.6 | 3.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.6 | 1.9 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.6 | 2.6 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.6 | 5.1 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.6 | 2.5 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.6 | 2.4 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.6 | 1.8 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.6 | 1.8 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.6 | 2.4 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.5 | 2.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.5 | 1.5 | GO:0034239 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.5 | 2.0 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.5 | 8.0 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.5 | 1.4 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.5 | 1.4 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.5 | 2.8 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.5 | 2.3 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.4 | 1.8 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.4 | 2.2 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.4 | 1.7 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.4 | 3.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.4 | 1.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.4 | 2.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.4 | 2.5 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.4 | 1.2 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.4 | 1.2 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.4 | 0.4 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.4 | 7.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.4 | 0.8 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.4 | 1.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.4 | 0.7 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.4 | 1.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.4 | 1.8 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.4 | 1.5 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.4 | 5.0 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.4 | 2.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.4 | 2.5 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.4 | 1.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.4 | 1.4 | GO:0072299 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.4 | 2.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.3 | 4.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 1.0 | GO:1901253 | negative regulation of dendritic cell cytokine production(GO:0002731) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.3 | 1.7 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.3 | 7.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 6.5 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.3 | 1.0 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.3 | 1.0 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.3 | 0.9 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.3 | 5.0 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.3 | 2.8 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.3 | 1.8 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.3 | 0.6 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.3 | 32.5 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.3 | 1.8 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.3 | 1.2 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.3 | 0.9 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.3 | 1.8 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.3 | 0.9 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.3 | 2.0 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.3 | 0.3 | GO:0042223 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) |
| 0.3 | 1.7 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.3 | 0.8 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.3 | 0.6 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.3 | 2.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.3 | 0.6 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.3 | 2.7 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.3 | 1.9 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.3 | 0.8 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.3 | 1.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 | 1.1 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.3 | 21.3 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.3 | 3.4 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.3 | 3.4 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.3 | 0.5 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 5.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.3 | 1.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.3 | 0.8 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.3 | 2.8 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.3 | 2.0 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.3 | 0.8 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.3 | 0.8 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.2 | 1.0 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.2 | 0.5 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.2 | 2.7 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.2 | 2.9 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 0.5 | GO:2000974 | negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.2 | 0.2 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.2 | 0.2 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.2 | 0.5 | GO:2000544 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.2 | 1.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 0.5 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.2 | 0.7 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.2 | 2.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 0.5 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.2 | 0.5 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.2 | 2.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 8.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 0.9 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.2 | 3.0 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.2 | 1.1 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.2 | 0.9 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.2 | 1.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.2 | 2.5 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.2 | 0.7 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.2 | 0.2 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.2 | 2.0 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.2 | 1.3 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.2 | 7.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.2 | 0.6 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.2 | 1.0 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.2 | 0.4 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.2 | 0.8 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.2 | 0.6 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.2 | 2.1 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.2 | 2.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 1.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 1.0 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.2 | 0.6 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.2 | 0.6 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.2 | 0.6 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 | 0.8 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.2 | 0.6 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.2 | 1.0 | GO:0043091 | response to herbicide(GO:0009635) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.2 | 0.8 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.2 | 0.4 | GO:0030473 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear migration along microfilament(GO:0031022) |
| 0.2 | 0.2 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.2 | 0.9 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 0.6 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.2 | 0.7 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.2 | 0.9 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.2 | 0.6 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.2 | 0.6 | GO:2000861 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) positive regulation of estrogen secretion(GO:2000863) regulation of estradiol secretion(GO:2000864) positive regulation of estradiol secretion(GO:2000866) |
| 0.2 | 0.4 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.2 | 0.9 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.2 | 0.9 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.2 | 0.5 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.2 | 0.7 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.2 | 0.4 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.2 | 0.4 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.2 | 0.2 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.2 | 3.4 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.2 | 0.5 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.2 | 3.1 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.2 | 2.1 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.2 | 6.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 1.4 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 1.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 2.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 0.8 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.2 | 1.9 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.2 | 0.6 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.2 | 0.6 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.2 | 0.8 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.2 | 0.2 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.2 | 0.5 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.2 | 2.8 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.2 | 1.5 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 0.6 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.2 | 0.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.4 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.1 | 2.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.9 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 3.0 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.1 | 0.9 | GO:0051661 | maintenance of centrosome location(GO:0051661) maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.9 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.1 | 0.7 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.1 | 1.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 1.3 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.6 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.1 | 0.6 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 3.5 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.3 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 2.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.6 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 2.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 1.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 1.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 10.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.4 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.5 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 2.0 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.1 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.1 | 0.4 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.1 | 2.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.8 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.7 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 2.5 | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) |
| 0.1 | 0.8 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.1 | 0.4 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.8 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 0.3 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 0.3 | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.1 | 0.4 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 2.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.5 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.6 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.4 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.3 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 | 1.0 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 0.3 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 0.5 | GO:0042746 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.1 | 6.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.7 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.4 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 | 0.4 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 1.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 3.0 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.7 | GO:0015853 | adenine transport(GO:0015853) |
| 0.1 | 0.6 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 2.1 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 0.2 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 0.7 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.1 | 1.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.8 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.4 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 3.4 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 0.8 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.1 | 0.5 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.7 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.6 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 1.3 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.8 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.7 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.7 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.8 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 1.2 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.1 | 1.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 1.0 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.1 | 2.6 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 3.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 1.7 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.6 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.7 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 9.6 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.6 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.1 | 0.4 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 0.4 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.1 | 0.5 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.7 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.1 | 3.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.5 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 0.3 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 0.4 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) |
| 0.1 | 0.3 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.1 | 0.7 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 0.4 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.7 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 1.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 1.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.3 | GO:0051292 | nuclear pore organization(GO:0006999) nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.1 | 3.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) mitotic G2/M transition checkpoint(GO:0044818) |
| 0.1 | 1.8 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.4 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.5 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 0.5 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 | 0.3 | GO:0009820 | alkaloid metabolic process(GO:0009820) neuroblast differentiation(GO:0014016) |
| 0.1 | 0.6 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.5 | GO:0048858 | cell part morphogenesis(GO:0032990) cell projection morphogenesis(GO:0048858) |
| 0.1 | 0.5 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.1 | 2.6 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 | 0.8 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.1 | 1.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.4 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.1 | 1.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.3 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.4 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 0.7 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.2 | GO:0021554 | cranial nerve development(GO:0021545) optic nerve development(GO:0021554) |
| 0.1 | 0.8 | GO:1904948 | midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.1 | 0.7 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 13.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.6 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) |
| 0.1 | 0.4 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 0.6 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.1 | 0.6 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 0.7 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 1.1 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.1 | 0.5 | GO:0035900 | response to isolation stress(GO:0035900) regulation of eating behavior(GO:1903998) |
| 0.1 | 0.3 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 2.2 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.1 | 0.5 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 0.3 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.1 | 1.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 9.9 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 0.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.4 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.1 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 0.8 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.1 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 1.2 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 0.3 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 0.5 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 1.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.1 | 0.4 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.7 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 0.6 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 7.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.2 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 0.2 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 0.2 | GO:0034182 | regulation of maintenance of sister chromatid cohesion(GO:0034091) regulation of maintenance of mitotic sister chromatid cohesion(GO:0034182) |
| 0.1 | 0.2 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 2.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 7.8 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 0.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.1 | 3.2 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.1 | 0.6 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.9 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.6 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.1 | 0.3 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 1.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.4 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 0.3 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 1.0 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 0.7 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.2 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 | 0.3 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 0.4 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 1.4 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 | 0.9 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.5 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.5 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.1 | 0.7 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.3 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) cranial skeletal system development(GO:1904888) |
| 0.1 | 0.5 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.3 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 0.1 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 1.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.7 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 0.6 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 0.8 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 1.0 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 0.1 | GO:0090024 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 1.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 1.8 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.1 | 0.2 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 | 0.6 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.3 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.9 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 1.3 | GO:0017145 | stem cell division(GO:0017145) |
| 0.1 | 1.2 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.1 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.2 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.3 | GO:0051402 | neuron apoptotic process(GO:0051402) |
| 0.1 | 0.6 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 0.2 | GO:0043247 | telomere maintenance in response to DNA damage(GO:0043247) |
| 0.1 | 0.3 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.4 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.1 | 1.1 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.6 | GO:0032648 | regulation of interferon-beta production(GO:0032648) |
| 0.1 | 1.1 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.2 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.1 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.3 | GO:0019050 | suppression by virus of host apoptotic process(GO:0019050) modulation by virus of host apoptotic process(GO:0039526) |
| 0.1 | 0.5 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.1 | 2.5 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.1 | GO:1904798 | regulation of core promoter binding(GO:1904796) positive regulation of core promoter binding(GO:1904798) |
| 0.1 | 1.0 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.1 | 0.1 | GO:1904044 | response to aldosterone(GO:1904044) |
| 0.1 | 0.4 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.1 | 0.7 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.9 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.3 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.3 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 3.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.0 | 0.3 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.6 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 1.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.4 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.0 | 1.0 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.1 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 1.0 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.4 | GO:0050715 | positive regulation of cytokine secretion(GO:0050715) |
| 0.0 | 0.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 1.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.7 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.3 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 1.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.7 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 1.2 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.2 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.9 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 2.6 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.4 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.4 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.2 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.6 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.3 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 1.3 | GO:0042044 | fluid transport(GO:0042044) |
| 0.0 | 8.6 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.5 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.5 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.4 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.0 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.3 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.9 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.6 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.2 | GO:1904683 | regulation of metalloendopeptidase activity(GO:1904683) negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.5 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.9 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.3 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 1.0 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.5 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.2 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.9 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 1.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.6 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 0.6 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.5 | GO:0031175 | neuron projection development(GO:0031175) |
| 0.0 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.7 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 1.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 2.6 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.4 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.2 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.9 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 0.0 | 0.7 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 1.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.7 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 2.2 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.1 | GO:0097479 | synaptic vesicle transport(GO:0048489) synaptic vesicle localization(GO:0097479) establishment of synaptic vesicle localization(GO:0097480) |
| 0.0 | 0.2 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.4 | GO:0006805 | xenobiotic metabolic process(GO:0006805) response to xenobiotic stimulus(GO:0009410) cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0001983 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 1.4 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 1.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:2000269 | fibroblast apoptotic process(GO:0044346) regulation of fibroblast apoptotic process(GO:2000269) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.5 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.0 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.2 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.2 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.0 | 0.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.4 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.0 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.5 | GO:0002092 | positive regulation of receptor internalization(GO:0002092) |
| 0.0 | 0.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.3 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.5 | GO:1903432 | regulation of TORC1 signaling(GO:1903432) |
| 0.0 | 0.1 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.0 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 | 0.1 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.2 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.7 | GO:0001942 | hair follicle development(GO:0001942) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.3 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.0 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 | 0.1 | GO:0045188 | optic nerve morphogenesis(GO:0021631) optic nerve structural organization(GO:0021633) regulation of circadian sleep/wake cycle, non-REM sleep(GO:0045188) |
| 0.0 | 1.0 | GO:0097202 | activation of cysteine-type endopeptidase activity(GO:0097202) |
| 0.0 | 0.3 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.1 | GO:0044854 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.6 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.2 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0001936 | regulation of endothelial cell proliferation(GO:0001936) |
| 0.0 | 0.1 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.0 | GO:0051866 | aggressive behavior(GO:0002118) general adaptation syndrome(GO:0051866) |
| 0.0 | 1.0 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.2 | GO:0001525 | angiogenesis(GO:0001525) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.3 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.4 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.1 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0006067 | ethanol metabolic process(GO:0006067) |
| 0.0 | 0.2 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.3 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.2 | GO:0051197 | positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 0.4 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.8 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.2 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.0 | 0.1 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.0 | 0.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.2 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.2 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.0 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 3.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 2.2 | 15.6 | GO:0002177 | manchette(GO:0002177) |
| 1.7 | 31.5 | GO:0036038 | MKS complex(GO:0036038) |
| 1.2 | 12.8 | GO:0036157 | outer dynein arm(GO:0036157) |
| 1.1 | 22.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.0 | 2.9 | GO:0001534 | radial spoke(GO:0001534) |
| 0.6 | 9.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.5 | 6.4 | GO:0097433 | dense body(GO:0097433) |
| 0.4 | 2.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 0.4 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.4 | 2.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.4 | 1.2 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.4 | 1.9 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.4 | 2.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.4 | 3.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 1.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 3.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 0.9 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.3 | 1.4 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.3 | 2.8 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.3 | 0.8 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.3 | 1.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.3 | 1.3 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.3 | 0.8 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.2 | 1.0 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.2 | 25.2 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.2 | 1.9 | GO:0042599 | lamellar body(GO:0042599) |
| 0.2 | 0.7 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 1.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 0.9 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.2 | 0.9 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.2 | 1.7 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 0.8 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.2 | 11.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 2.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 1.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 1.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 4.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 3.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 5.7 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 0.2 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.2 | 0.9 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.2 | 0.5 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.2 | 5.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 3.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 0.7 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 1.4 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 3.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 1.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 0.5 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.2 | 0.6 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.2 | 3.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 0.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 2.0 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 1.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 2.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 3.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 3.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 3.2 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.5 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.6 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 1.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.6 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 1.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.6 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 1.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 3.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 1.9 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 7.6 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 2.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.7 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 8.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.6 | GO:1990393 | Cul7-RING ubiquitin ligase complex(GO:0031467) 3M complex(GO:1990393) |
| 0.1 | 7.6 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 3.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.8 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 4.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.3 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.8 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.5 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.1 | 1.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 7.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.7 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.1 | 1.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.5 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 0.9 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.7 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.5 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.5 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.3 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 0.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.6 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.1 | 1.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 5.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.6 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 1.0 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.2 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.2 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 1.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 0.4 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 0.2 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 1.6 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 5.0 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 1.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 3.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.8 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 1.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 1.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 7.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 8.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.6 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 1.5 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 1.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.9 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 8.5 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 1.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.2 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.5 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.6 | GO:0071010 | prespliceosome(GO:0071010) |
| 0.0 | 3.4 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 8.5 | GO:0019867 | outer membrane(GO:0019867) |
| 0.0 | 0.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.7 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 3.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 11.8 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.5 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.0 | GO:0032280 | symmetric synapse(GO:0032280) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.8 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 23.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.0 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 4.5 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 13.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.9 | 7.8 | GO:0016160 | amylase activity(GO:0016160) |
| 1.7 | 8.6 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 1.5 | 6.0 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 1.3 | 6.6 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 1.2 | 24.4 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 1.1 | 7.8 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 1.1 | 3.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 1.1 | 6.3 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 1.0 | 6.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 1.0 | 3.0 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 1.0 | 4.9 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.8 | 11.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.8 | 3.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.7 | 6.4 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.7 | 2.1 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.7 | 9.2 | GO:0070330 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) aromatase activity(GO:0070330) |
| 0.6 | 1.9 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.6 | 1.8 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.6 | 4.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.6 | 0.6 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.6 | 11.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.5 | 2.7 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.5 | 9.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.5 | 1.6 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.5 | 2.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.5 | 1.5 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.5 | 3.6 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.5 | 1.5 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.5 | 10.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.5 | 2.8 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.4 | 0.9 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.4 | 1.3 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.4 | 1.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.4 | 2.9 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.4 | 1.6 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.4 | 1.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.4 | 2.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.4 | 8.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 2.8 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.4 | 1.6 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.4 | 7.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 1.9 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.4 | 1.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.4 | 1.5 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.4 | 3.4 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.4 | 0.7 | GO:0004096 | catalase activity(GO:0004096) |
| 0.4 | 1.5 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.4 | 1.4 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.4 | 0.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.4 | 2.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.4 | 1.4 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.4 | 1.8 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.3 | 2.4 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.3 | 2.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 1.0 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.3 | 4.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 8.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.3 | 2.8 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.3 | 2.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 0.3 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.3 | 0.9 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.3 | 0.9 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.3 | 1.7 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.3 | 0.9 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.3 | 1.4 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.3 | 1.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.3 | 0.8 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.3 | 1.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.3 | 1.9 | GO:0032934 | sterol binding(GO:0032934) |
| 0.3 | 3.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 16.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.3 | 0.8 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.3 | 0.8 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.2 | 0.7 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 1.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 2.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 1.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 0.2 | GO:0017076 | purine nucleotide binding(GO:0017076) |
| 0.2 | 1.4 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.2 | 1.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 3.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 0.9 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.2 | 0.9 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.2 | 5.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 0.6 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.2 | 5.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.2 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 2.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 0.6 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.2 | 0.6 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.2 | 11.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 0.6 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.2 | 0.8 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.2 | 4.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 5.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.2 | 0.6 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.2 | 3.7 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.2 | 0.9 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.2 | 1.8 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 2.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 0.5 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 1.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 2.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 1.5 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.2 | 1.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 1.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 1.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 1.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.2 | 30.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 0.5 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 0.6 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 1.2 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.2 | 1.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.4 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 1.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.7 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.4 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.4 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.6 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.8 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 1.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.7 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.4 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.5 | GO:0016301 | kinase activity(GO:0016301) |
| 0.1 | 2.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.4 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.1 | 1.8 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 3.9 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 0.9 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 1.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.5 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.9 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 5.1 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.4 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 6.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 2.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.5 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 0.5 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.1 | 0.5 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 3.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.0 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 1.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 4.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 2.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 3.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.4 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.4 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 4.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 4.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.2 | GO:0005503 | all-trans retinal binding(GO:0005503) |
| 0.1 | 1.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.4 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.1 | 0.5 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.5 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 1.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.1 | 1.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 3.3 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.1 | 0.3 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.1 | 0.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 2.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 8.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.7 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.3 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 5.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.5 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 0.5 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.1 | 0.4 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.4 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 1.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.1 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.4 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 0.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 2.9 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 0.8 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.6 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 1.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.6 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 0.8 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 0.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 2.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 1.8 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 2.0 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 1.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.2 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 0.2 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 2.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 0.8 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 3.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 1.9 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 0.1 | GO:0001083 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) |
| 0.1 | 0.9 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.2 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 2.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 0.3 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 0.2 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 0.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 1.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 0.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.6 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 1.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.9 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.3 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.2 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 1.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 2.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 2.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.2 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 0.2 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.1 | 0.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) potassium ion antiporter activity(GO:0022821) |
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 1.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.4 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.8 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.2 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 0.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 2.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 3.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.0 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.3 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 1.8 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.1 | 1.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.2 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.5 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0019863 | immunoglobulin receptor activity(GO:0019763) IgE binding(GO:0019863) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.5 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.3 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 1.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.3 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.2 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 3.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 1.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 1.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 4.9 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 1.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 27.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.4 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 6.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 3.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.0 | 0.5 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 2.1 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.7 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 1.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 2.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.4 | GO:0042166 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 2.0 | GO:0016773 | phosphotransferase activity, alcohol group as acceptor(GO:0016773) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 1.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.0 | 0.4 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.0 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.2 | 2.4 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 8.9 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 0.5 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 8.7 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.1 | 0.4 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.8 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.8 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.1 | 0.8 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 2.1 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.4 | PID_TRAIL_PATHWAY | TRAIL signaling pathway |
| 0.1 | 5.3 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.1 | 0.4 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 0.7 | PID_CD8_TCR_DOWNSTREAM_PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 3.7 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.5 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.4 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.2 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 2.9 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.0 | 0.5 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.9 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 2.8 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.2 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.6 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.0 | 3.5 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.2 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.5 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 1.0 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.7 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 2.3 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 2.8 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.7 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 5.3 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID_TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.8 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.3 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.0 | 0.1 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.8 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.1 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.1 | PID_ATM_PATHWAY | ATM pathway |
| 0.0 | 0.2 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 12.4 | REACTOME_DIGESTION_OF_DIETARY_CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 1.0 | 13.7 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.8 | 13.6 | REACTOME_XENOBIOTICS | Genes involved in Xenobiotics |
| 0.7 | 20.9 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.6 | 14.5 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.4 | 4.3 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_24_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 8.5 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 11.5 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 0.6 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 2.2 | REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 2.5 | REACTOME_ANDROGEN_BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 3.5 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.1 | REACTOME_APOBEC3G_MEDIATED_RESISTANCE_TO_HIV1_INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 2.0 | REACTOME_REGULATED_PROTEOLYSIS_OF_P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 5.1 | REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.2 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 4.5 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.7 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.6 | REACTOME_RECYCLING_OF_BILE_ACIDS_AND_SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.3 | REACTOME_STEROID_HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.3 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_LATE_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 3.5 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 2.1 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.3 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.4 | REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 0.3 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.3 | REACTOME_BINDING_AND_ENTRY_OF_HIV_VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 2.2 | REACTOME_COMPLEMENT_CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.9 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.8 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 2.0 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.2 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 0.8 | REACTOME_RAF_MAP_KINASE_CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 1.0 | REACTOME_SEROTONIN_RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 5.8 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.5 | REACTOME_COMMON_PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.1 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 2.7 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.6 | REACTOME_ABACAVIR_TRANSPORT_AND_METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.3 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.1 | REACTOME_THROMBIN_SIGNALLING_THROUGH_PROTEINASE_ACTIVATED_RECEPTORS_PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 2.4 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.1 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.4 | REACTOME_INTERACTIONS_OF_VPR_WITH_HOST_CELLULAR_PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.9 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.9 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.4 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME_INTEGRATION_OF_PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.4 | REACTOME_ENDOSOMAL_VACUOLAR_PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.8 | REACTOME_MICRORNA_MIRNA_BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.2 | REACTOME_CROSS_PRESENTATION_OF_SOLUBLE_EXOGENOUS_ANTIGENS_ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.9 | REACTOME_TETRAHYDROBIOPTERIN_BH4_SYNTHESIS_RECYCLING_SALVAGE_AND_REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.7 | REACTOME_ACTIVATED_TAK1_MEDIATES_P38_MAPK_ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 1.0 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 16.4 | REACTOME_GENERIC_TRANSCRIPTION_PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.3 | REACTOME_TRANSPORT_OF_MATURE_TRANSCRIPT_TO_CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 1.1 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.7 | REACTOME_DOWNSTREAM_TCR_SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.5 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.7 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 2.2 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.7 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.0 | 0.5 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.9 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME_SOS_MEDIATED_SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.4 | REACTOME_FORMATION_OF_THE_HIV1_EARLY_ELONGATION_COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_7ALPHA_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.1 | REACTOME_IRON_UPTAKE_AND_TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.0 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.9 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.1 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.5 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.1 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.5 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME_TAK1_ACTIVATES_NFKB_BY_PHOSPHORYLATION_AND_ACTIVATION_OF_IKKS_COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.1 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.0 | REACTOME_INHIBITION_OF_REPLICATION_INITIATION_OF_DAMAGED_DNA_BY_RB1_E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |