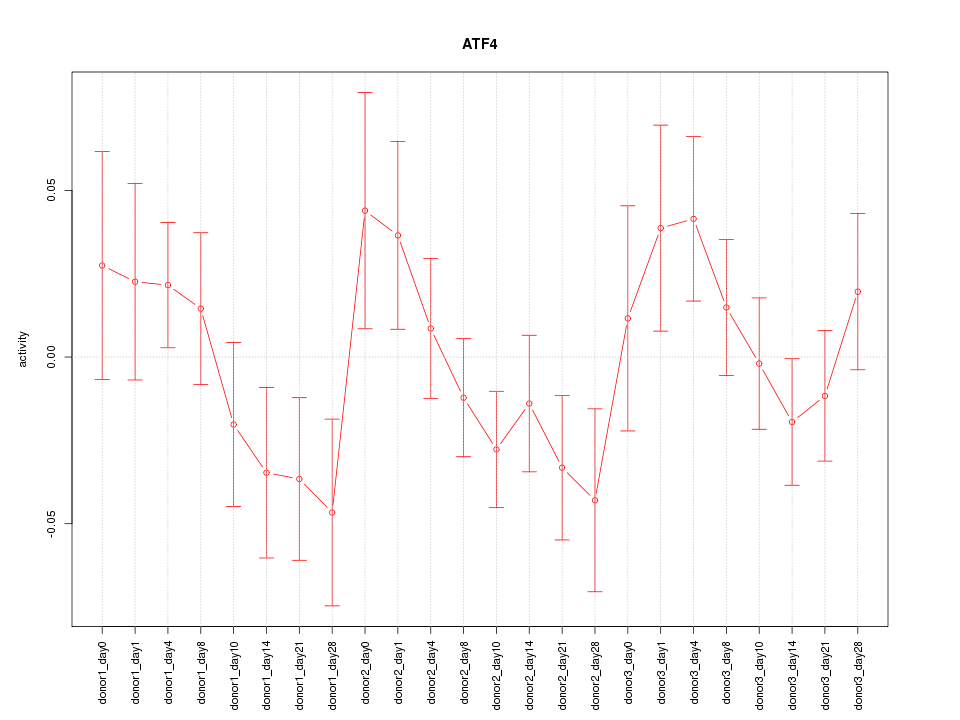
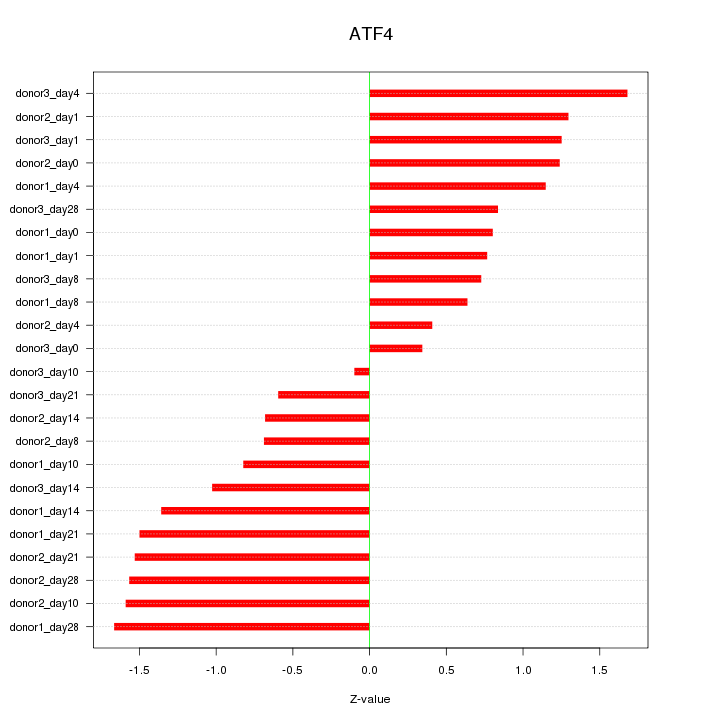
Motif ID: ATF4
Z-value: 1.105
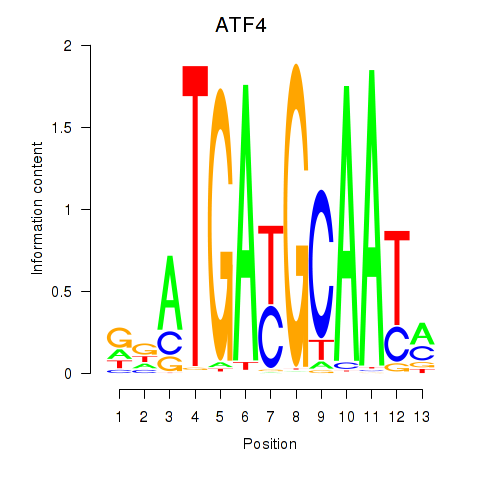
Transcription factors associated with ATF4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| ATF4 | ENSG00000128272.10 | ATF4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF4 | hg19_v2_chr22_+_39916558_39916579 | 0.68 | 2.5e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 1.7 | 5.2 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 1.6 | 9.6 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 1.0 | 7.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.9 | 2.8 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.9 | 3.6 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.6 | 3.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.6 | 2.5 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.5 | 1.6 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.4 | 2.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.4 | 1.7 | GO:0042853 | L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.4 | 1.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.4 | 2.0 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 2.7 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.3 | 1.7 | GO:0060356 | leucine import(GO:0060356) |
| 0.3 | 1.3 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.3 | 1.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.3 | 1.8 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.3 | 1.3 | GO:1904640 | response to methionine(GO:1904640) |
| 0.2 | 1.0 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.2 | 1.0 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.2 | 0.6 | GO:0050894 | determination of affect(GO:0050894) |
| 0.2 | 0.6 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.2 | 1.1 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.2 | 0.5 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.2 | 0.5 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.2 | 0.9 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.4 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.1 | 2.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 1.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.4 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.1 | 0.5 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.3 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.1 | 1.4 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.1 | 0.7 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 1.4 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.3 | GO:2000639 | negative regulation of triglyceride biosynthetic process(GO:0010868) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.4 | GO:1903912 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.9 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 2.8 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.0 | 0.8 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 1.0 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.6 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.6 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.4 | GO:0030163 | protein catabolic process(GO:0030163) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.4 | 5.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 0.8 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.0 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 1.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 1.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 2.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 3.1 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 10.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.6 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 2.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0050421 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 1.6 | 9.6 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 1.0 | 7.1 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.9 | 2.7 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.9 | 3.6 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.8 | 2.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.7 | 2.0 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.6 | 3.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.6 | 2.5 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.6 | 2.8 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.5 | 1.6 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.3 | 1.0 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.3 | 1.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.3 | 1.8 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 0.6 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.2 | 0.6 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.2 | 0.5 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.2 | 0.9 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.4 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.1 | 1.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 2.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 5.4 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 1.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.5 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 1.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 1.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 3.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.6 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 2.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.4 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 8.9 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.6 | ST_INTERLEUKIN_4_PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 2.7 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.9 | PID_IL2_1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.7 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.0 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.0 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.0 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.8 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.3 | PID_MYC_PATHWAY | C-MYC pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.6 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 8.9 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 5.2 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 5.4 | REACTOME_GRB2_SOS_PROVIDES_LINKAGE_TO_MAPK_SIGNALING_FOR_INTERGRINS_ | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 5.2 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.4 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.2 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.0 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 3.3 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 3.7 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 2.0 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.9 | REACTOME_GROWTH_HORMONE_RECEPTOR_SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 2.8 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.6 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |