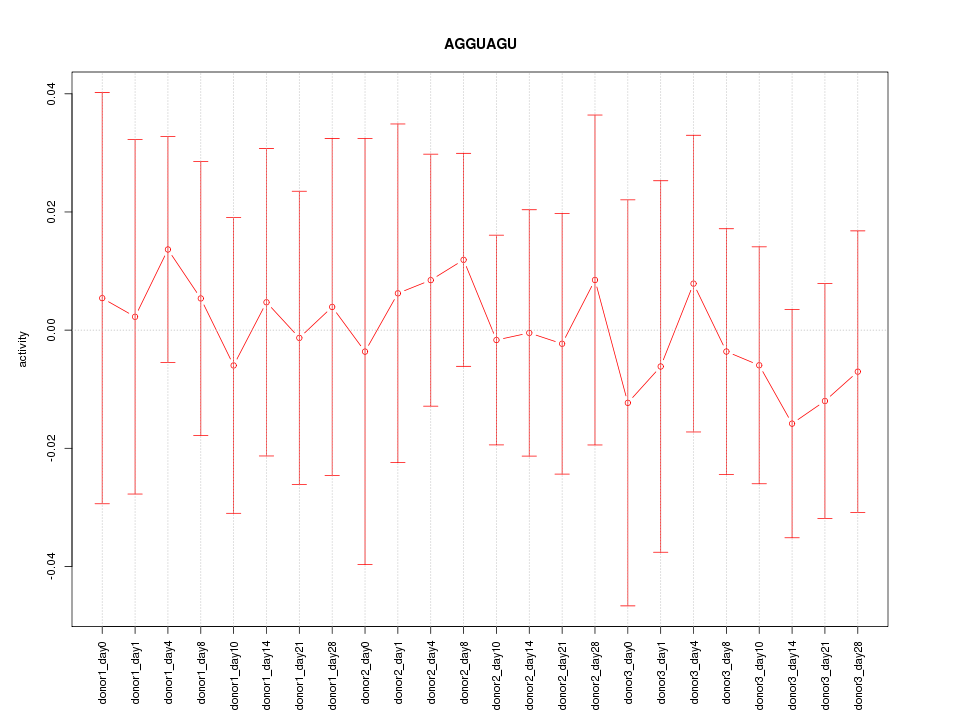
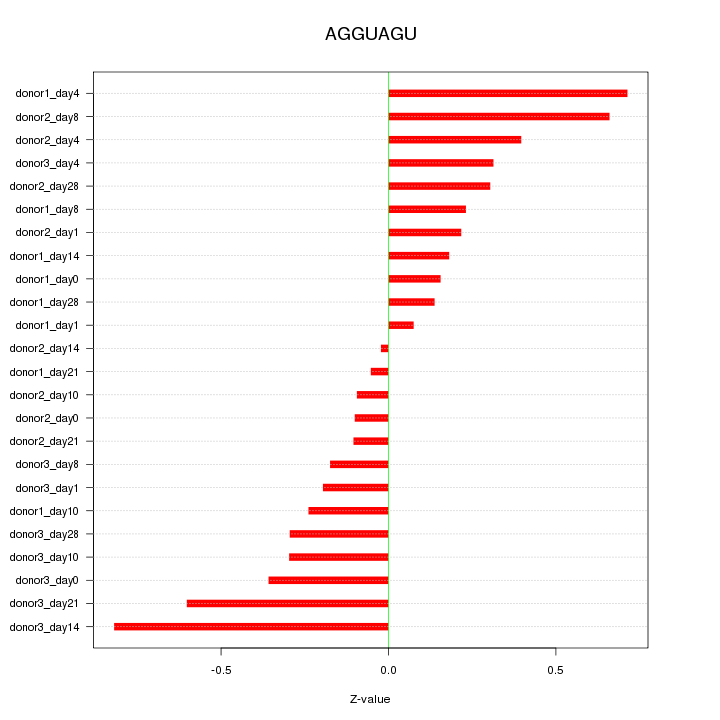
|
chr17_-_48278983
|
0.281
|
ENST00000225964.5
|
COL1A1
|
collagen, type I, alpha 1
|
|
chrX_-_132549506
|
0.269
|
ENST00000370828.3
|
GPC4
|
glypican 4
|
|
chr7_+_94285637
|
0.242
|
ENST00000482108.1
ENST00000488574.1
|
PEG10
|
paternally expressed 10
|
|
chr7_-_75368248
|
0.242
|
ENST00000434438.2
ENST00000336926.6
|
HIP1
|
huntingtin interacting protein 1
|
|
chr19_-_47975417
|
0.240
|
ENST00000236877.6
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2
|
|
chr11_+_63580855
|
0.203
|
ENST00000294244.4
|
C11orf84
|
chromosome 11 open reading frame 84
|
|
chr3_+_158991025
|
0.198
|
ENST00000337808.6
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough
|
|
chr8_+_92082424
|
0.197
|
ENST00000285420.4
ENST00000404789.3
|
OTUD6B
|
OTU domain containing 6B
|
|
chr3_+_159557637
|
0.193
|
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr12_+_12870055
|
0.165
|
ENST00000228872.4
|
CDKN1B
|
cyclin-dependent kinase inhibitor 1B (p27, Kip1)
|
|
chr12_-_59313270
|
0.160
|
ENST00000379141.4
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr3_-_120003941
|
0.156
|
ENST00000464295.1
|
GPR156
|
G protein-coupled receptor 156
|
|
chr18_-_24445729
|
0.152
|
ENST00000383168.4
|
AQP4
|
aquaporin 4
|
|
chr8_+_81397876
|
0.147
|
ENST00000430430.1
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr18_-_74207146
|
0.146
|
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516
|
|
chr20_+_37434329
|
0.141
|
ENST00000299824.1
ENST00000373331.2
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|
|
chr4_+_56212270
|
0.140
|
ENST00000264228.4
|
SRD5A3
|
steroid 5 alpha-reductase 3
|
|
chr6_+_105404899
|
0.138
|
ENST00000345080.4
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr9_-_125693757
|
0.135
|
ENST00000373656.3
|
ZBTB26
|
zinc finger and BTB domain containing 26
|
|
chr17_+_55333876
|
0.119
|
ENST00000284073.2
|
MSI2
|
musashi RNA-binding protein 2
|
|
chr16_+_22217577
|
0.109
|
ENST00000263026.5
|
EEF2K
|
eukaryotic elongation factor-2 kinase
|
|
chrX_+_95939711
|
0.108
|
ENST00000373049.4
ENST00000324765.8
|
DIAPH2
|
diaphanous-related formin 2
|
|
chr2_+_204192942
|
0.106
|
ENST00000295851.5
ENST00000261017.5
|
ABI2
|
abl-interactor 2
|
|
chr3_+_110790590
|
0.104
|
ENST00000485303.1
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr17_+_8213590
|
0.100
|
ENST00000361926.3
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|
|
chr5_+_153825510
|
0.098
|
ENST00000297109.6
|
SAP30L
|
SAP30-like
|
|
chr9_+_4662282
|
0.098
|
ENST00000381883.2
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2
|
|
chr20_+_49348081
|
0.098
|
ENST00000371610.2
|
PARD6B
|
par-6 family cell polarity regulator beta
|
|
chr5_+_49961727
|
0.097
|
ENST00000505697.2
ENST00000503750.2
ENST00000514342.2
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr22_+_50781723
|
0.095
|
ENST00000359139.3
ENST00000395741.3
ENST00000395744.3
|
PPP6R2
|
protein phosphatase 6, regulatory subunit 2
|
|
chr7_+_94023873
|
0.094
|
ENST00000297268.6
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr12_+_66217911
|
0.092
|
ENST00000403681.2
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr1_+_65210772
|
0.088
|
ENST00000371072.4
ENST00000294428.3
|
RAVER2
|
ribonucleoprotein, PTB-binding 2
|
|
chr18_+_19749386
|
0.087
|
ENST00000269216.3
|
GATA6
|
GATA binding protein 6
|
|
chr16_+_29789561
|
0.085
|
ENST00000400752.4
|
ZG16
|
zymogen granule protein 16
|
|
chr9_+_109625378
|
0.083
|
ENST00000277225.5
ENST00000457913.1
ENST00000472574.1
|
ZNF462
|
zinc finger protein 462
|
|
chr1_-_204380919
|
0.083
|
ENST00000367188.4
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B
|
|
chr12_+_54402790
|
0.083
|
ENST00000040584.4
|
HOXC8
|
homeobox C8
|
|
chr5_+_172068232
|
0.082
|
ENST00000520919.1
ENST00000522853.1
ENST00000369800.5
|
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B
|
|
chr16_+_70332956
|
0.081
|
ENST00000288071.6
ENST00000393657.2
ENST00000355992.3
ENST00000567706.1
|
DDX19B
RP11-529K1.3
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19B
Uncharacterized protein
|
|
chr4_+_184826418
|
0.081
|
ENST00000308497.4
ENST00000438269.1
|
STOX2
|
storkhead box 2
|
|
chr6_-_134373732
|
0.080
|
ENST00000275230.5
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12
|
|
chr20_+_277737
|
0.078
|
ENST00000382352.3
|
ZCCHC3
|
zinc finger, CCHC domain containing 3
|
|
chr4_+_113066552
|
0.076
|
ENST00000309733.5
|
C4orf32
|
chromosome 4 open reading frame 32
|
|
chr8_-_82754427
|
0.075
|
ENST00000353788.4
ENST00000520618.1
ENST00000518183.1
ENST00000396330.2
ENST00000519119.1
ENST00000345957.4
|
SNX16
|
sorting nexin 16
|
|
chr19_-_51054299
|
0.075
|
ENST00000599957.1
|
LRRC4B
|
leucine rich repeat containing 4B
|
|
chr1_+_26737253
|
0.071
|
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr17_+_2699697
|
0.069
|
ENST00000254695.8
ENST00000366401.4
ENST00000542807.1
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chrX_+_118108571
|
0.068
|
ENST00000304778.7
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr6_-_42419649
|
0.068
|
ENST00000372922.4
ENST00000541110.1
ENST00000372917.4
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr21_+_30671189
|
0.064
|
ENST00000286800.3
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr17_-_46682321
|
0.064
|
ENST00000225648.3
ENST00000484302.2
|
HOXB6
|
homeobox B6
|
|
chr2_+_170590321
|
0.063
|
ENST00000392647.2
|
KLHL23
|
kelch-like family member 23
|
|
chr2_+_32390925
|
0.063
|
ENST00000440718.1
ENST00000379343.2
ENST00000282587.5
ENST00000435660.1
ENST00000538303.1
ENST00000357055.3
ENST00000406369.1
|
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6
|
|
chr8_+_58907104
|
0.063
|
ENST00000361488.3
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr12_+_118814344
|
0.062
|
ENST00000397564.2
|
SUDS3
|
suppressor of defective silencing 3 homolog (S. cerevisiae)
|
|
chr20_+_306221
|
0.061
|
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12
|
|
chr8_+_22102626
|
0.060
|
ENST00000519237.1
ENST00000397802.4
|
POLR3D
|
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa
|
|
chr16_-_30621663
|
0.060
|
ENST00000287461.3
|
ZNF689
|
zinc finger protein 689
|
|
chr19_+_14063278
|
0.059
|
ENST00000254337.6
|
DCAF15
|
DDB1 and CUL4 associated factor 15
|
|
chr4_-_18023350
|
0.059
|
ENST00000539056.1
ENST00000382226.5
ENST00000326877.4
|
LCORL
|
ligand dependent nuclear receptor corepressor-like
|
|
chrX_-_24690771
|
0.058
|
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr1_-_231473578
|
0.057
|
ENST00000360394.2
ENST00000366645.1
|
EXOC8
|
exocyst complex component 8
|
|
chr16_-_89007491
|
0.057
|
ENST00000327483.5
ENST00000564416.1
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr15_-_38856836
|
0.057
|
ENST00000450598.2
ENST00000559830.1
ENST00000558164.1
ENST00000310803.5
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated)
|
|
chr2_-_128615681
|
0.056
|
ENST00000409955.1
ENST00000272645.4
|
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D
|
|
chr2_+_198570081
|
0.053
|
ENST00000282276.6
|
MARS2
|
methionyl-tRNA synthetase 2, mitochondrial
|
|
chr3_-_21792838
|
0.052
|
ENST00000281523.2
|
ZNF385D
|
zinc finger protein 385D
|
|
chr1_-_92351769
|
0.051
|
ENST00000212355.4
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr10_-_38146510
|
0.050
|
ENST00000395867.3
|
ZNF248
|
zinc finger protein 248
|
|
chr2_+_232260254
|
0.050
|
ENST00000287590.5
|
B3GNT7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7
|
|
chr11_+_7273181
|
0.050
|
ENST00000318881.6
|
SYT9
|
synaptotagmin IX
|
|
chr12_-_57400227
|
0.050
|
ENST00000300101.2
|
ZBTB39
|
zinc finger and BTB domain containing 39
|
|
chr5_-_35230771
|
0.049
|
ENST00000342362.5
|
PRLR
|
prolactin receptor
|
|
chr20_-_5591626
|
0.049
|
ENST00000379019.4
|
GPCPD1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae)
|
|
chrX_-_129244655
|
0.049
|
ENST00000335997.7
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr6_-_99797522
|
0.049
|
ENST00000389677.5
|
FAXC
|
failed axon connections homolog (Drosophila)
|
|
chr1_+_60280458
|
0.048
|
ENST00000455990.1
ENST00000371208.3
|
HOOK1
|
hook microtubule-tethering protein 1
|
|
chr5_-_37371278
|
0.047
|
ENST00000231498.3
|
NUP155
|
nucleoporin 155kDa
|
|
chrX_-_83757399
|
0.047
|
ENST00000373177.2
ENST00000297977.5
ENST00000506585.2
ENST00000449553.2
|
HDX
|
highly divergent homeobox
|
|
chrX_+_2746850
|
0.047
|
ENST00000381163.3
ENST00000338623.5
ENST00000542787.1
|
GYG2
|
glycogenin 2
|
|
chr7_-_83824169
|
0.046
|
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr7_+_77325738
|
0.046
|
ENST00000334955.8
|
RSBN1L
|
round spermatid basic protein 1-like
|
|
chr5_-_39074479
|
0.044
|
ENST00000514735.1
ENST00000296782.5
ENST00000357387.3
|
RICTOR
|
RPTOR independent companion of MTOR, complex 2
|
|
chr15_-_34502278
|
0.043
|
ENST00000559515.1
ENST00000256544.3
ENST00000560108.1
ENST00000559462.1
|
KATNBL1
|
katanin p80 subunit B-like 1
|
|
chr11_-_796197
|
0.041
|
ENST00000530360.1
ENST00000528606.1
ENST00000320230.5
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr4_+_17616253
|
0.038
|
ENST00000237380.7
|
MED28
|
mediator complex subunit 28
|
|
chr18_+_55711575
|
0.036
|
ENST00000356462.6
ENST00000400345.3
ENST00000589054.1
ENST00000256832.7
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase
|
|
chr1_+_193091080
|
0.036
|
ENST00000367435.3
|
CDC73
|
cell division cycle 73
|
|
chr16_+_70380732
|
0.035
|
ENST00000302243.7
|
DDX19A
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19A
|
|
chr13_-_103053946
|
0.033
|
ENST00000376131.4
|
FGF14
|
fibroblast growth factor 14
|
|
chr1_-_17766198
|
0.033
|
ENST00000375436.4
|
RCC2
|
regulator of chromosome condensation 2
|
|
chr7_+_18535346
|
0.032
|
ENST00000405010.3
ENST00000406451.4
ENST00000428307.2
|
HDAC9
|
histone deacetylase 9
|
|
chr5_-_131347306
|
0.032
|
ENST00000296869.4
ENST00000379249.3
ENST00000379272.2
ENST00000379264.2
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr7_+_114055052
|
0.031
|
ENST00000462331.1
ENST00000408937.3
ENST00000403559.4
ENST00000350908.4
ENST00000393498.2
ENST00000393495.3
ENST00000378237.3
ENST00000393489.3
|
FOXP2
|
forkhead box P2
|
|
chr7_+_104654623
|
0.031
|
ENST00000311117.3
ENST00000334877.4
ENST00000257745.4
ENST00000334914.7
ENST00000478990.1
ENST00000495267.1
ENST00000476671.1
|
KMT2E
|
lysine (K)-specific methyltransferase 2E
|
|
chr12_-_498620
|
0.031
|
ENST00000399788.2
ENST00000382815.4
|
KDM5A
|
lysine (K)-specific demethylase 5A
|
|
chr6_-_35464727
|
0.030
|
ENST00000402886.3
|
TEAD3
|
TEA domain family member 3
|
|
chr6_+_134274322
|
0.029
|
ENST00000367871.1
ENST00000237264.4
|
TBPL1
|
TBP-like 1
|
|
chr3_+_61547585
|
0.029
|
ENST00000295874.10
ENST00000474889.1
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr12_-_25102252
|
0.029
|
ENST00000261192.7
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr9_+_133710453
|
0.028
|
ENST00000318560.5
|
ABL1
|
c-abl oncogene 1, non-receptor tyrosine kinase
|
|
chr17_+_68165657
|
0.027
|
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr3_-_28390581
|
0.027
|
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2
|
|
chr9_+_128509624
|
0.026
|
ENST00000342287.5
ENST00000373487.4
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chrX_-_24045303
|
0.026
|
ENST00000328046.8
|
KLHL15
|
kelch-like family member 15
|
|
chr20_-_31071239
|
0.026
|
ENST00000359676.5
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr3_-_69435224
|
0.025
|
ENST00000398540.3
|
FRMD4B
|
FERM domain containing 4B
|
|
chr6_+_133562472
|
0.025
|
ENST00000430974.2
ENST00000367895.5
ENST00000355167.3
ENST00000355286.6
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr5_+_38846101
|
0.025
|
ENST00000274276.3
|
OSMR
|
oncostatin M receptor
|
|
chr10_+_98592009
|
0.024
|
ENST00000540664.1
ENST00000371103.3
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr2_-_39664405
|
0.023
|
ENST00000341681.5
ENST00000263881.3
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr20_+_3869423
|
0.022
|
ENST00000497424.1
|
PANK2
|
pantothenate kinase 2
|
|
chr17_-_71228357
|
0.021
|
ENST00000583024.1
ENST00000403627.3
ENST00000405159.3
ENST00000581110.1
|
FAM104A
|
family with sequence similarity 104, member A
|
|
chr16_-_23160591
|
0.021
|
ENST00000219689.7
|
USP31
|
ubiquitin specific peptidase 31
|
|
chr5_-_158526756
|
0.021
|
ENST00000313708.6
ENST00000517373.1
|
EBF1
|
early B-cell factor 1
|
|
chr21_+_42539701
|
0.020
|
ENST00000330333.6
ENST00000328735.6
ENST00000347667.5
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr9_-_111929560
|
0.020
|
ENST00000561981.2
|
FRRS1L
|
ferric-chelate reductase 1-like
|
|
chr6_+_4890226
|
0.020
|
ENST00000343762.5
|
CDYL
|
chromodomain protein, Y-like
|
|
chr1_-_179198702
|
0.020
|
ENST00000502732.1
|
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase
|
|
chr12_-_76478686
|
0.019
|
ENST00000261182.8
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr12_-_77459306
|
0.019
|
ENST00000547316.1
ENST00000416496.2
ENST00000550669.1
ENST00000322886.7
|
E2F7
|
E2F transcription factor 7
|
|
chr5_-_131132614
|
0.019
|
ENST00000307968.7
ENST00000307954.8
|
FNIP1
|
folliculin interacting protein 1
|
|
chr9_+_115983808
|
0.018
|
ENST00000374210.6
ENST00000374212.4
|
SLC31A1
|
solute carrier family 31 (copper transporter), member 1
|
|
chr15_+_36887069
|
0.018
|
ENST00000566807.1
ENST00000567389.1
ENST00000562877.1
|
C15orf41
|
chromosome 15 open reading frame 41
|
|
chr1_-_51984908
|
0.018
|
ENST00000371730.2
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr15_+_90544532
|
0.015
|
ENST00000268154.4
|
ZNF710
|
zinc finger protein 710
|
|
chr9_-_79520989
|
0.015
|
ENST00000376713.3
ENST00000376718.3
ENST00000428286.1
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr1_-_200379180
|
0.015
|
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281
|
|
chr17_+_30771279
|
0.015
|
ENST00000261712.3
ENST00000578213.1
ENST00000457654.2
ENST00000579451.1
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr4_+_20702059
|
0.015
|
ENST00000444671.2
ENST00000510700.1
ENST00000506745.1
ENST00000514663.1
ENST00000509469.1
ENST00000515339.1
ENST00000513861.1
ENST00000502374.1
ENST00000538990.1
ENST00000511160.1
ENST00000504630.1
ENST00000513590.1
ENST00000514292.1
ENST00000502938.1
ENST00000509625.1
ENST00000505160.1
ENST00000507634.1
ENST00000513459.1
ENST00000511089.1
|
PACRGL
|
PARK2 co-regulated-like
|
|
chr19_+_531713
|
0.015
|
ENST00000215574.4
|
CDC34
|
cell division cycle 34
|
|
chr22_+_48972118
|
0.015
|
ENST00000358295.5
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5
|
|
chr18_+_67068228
|
0.015
|
ENST00000382713.5
|
DOK6
|
docking protein 6
|
|
chr3_+_49209023
|
0.014
|
ENST00000332780.2
|
KLHDC8B
|
kelch domain containing 8B
|
|
chrX_+_17393543
|
0.013
|
ENST00000380060.3
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr9_+_6413317
|
0.012
|
ENST00000276893.5
ENST00000381373.3
|
UHRF2
|
ubiquitin-like with PHD and ring finger domains 2, E3 ubiquitin protein ligase
|
|
chr2_+_179345173
|
0.011
|
ENST00000234453.5
|
PLEKHA3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3
|
|
chr10_+_97803151
|
0.011
|
ENST00000403870.3
ENST00000265992.5
ENST00000465148.2
ENST00000534974.1
|
CCNJ
|
cyclin J
|
|
chr17_-_46608272
|
0.011
|
ENST00000577092.1
ENST00000239174.6
|
HOXB1
|
homeobox B1
|
|
chr12_-_69326940
|
0.011
|
ENST00000549781.1
ENST00000548262.1
ENST00000551568.1
ENST00000548954.1
|
CPM
|
carboxypeptidase M
|
|
chr12_+_68042495
|
0.011
|
ENST00000344096.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2
|
|
chr1_+_109102652
|
0.011
|
ENST00000370035.3
ENST00000405454.1
|
FAM102B
|
family with sequence similarity 102, member B
|
|
chr6_+_24403144
|
0.010
|
ENST00000274747.7
ENST00000543597.1
ENST00000535061.1
ENST00000378353.1
ENST00000378386.3
ENST00000443868.2
|
MRS2
|
MRS2 magnesium transporter
|
|
chr2_-_50574856
|
0.010
|
ENST00000342183.5
|
NRXN1
|
neurexin 1
|
|
chr14_+_58666824
|
0.009
|
ENST00000254286.4
|
ACTR10
|
actin-related protein 10 homolog (S. cerevisiae)
|
|
chr8_+_37594130
|
0.009
|
ENST00000518526.1
ENST00000523887.1
ENST00000276461.5
|
ERLIN2
|
ER lipid raft associated 2
|
|
chr5_-_74326724
|
0.009
|
ENST00000322348.4
|
GCNT4
|
glucosaminyl (N-acetyl) transferase 4, core 2
|
|
chr11_+_111945011
|
0.009
|
ENST00000532163.1
ENST00000280352.9
ENST00000530104.1
ENST00000526879.1
ENST00000393047.3
ENST00000525785.1
|
C11orf57
|
chromosome 11 open reading frame 57
|
|
chr3_-_150264272
|
0.009
|
ENST00000491660.1
ENST00000487153.1
ENST00000239944.2
|
SERP1
|
stress-associated endoplasmic reticulum protein 1
|
|
chr2_+_242641442
|
0.008
|
ENST00000313552.6
ENST00000406941.1
|
ING5
|
inhibitor of growth family, member 5
|
|
chr12_-_123450986
|
0.008
|
ENST00000344275.7
ENST00000442833.2
ENST00000280560.8
ENST00000540285.1
ENST00000346530.5
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9
|
|
chr17_+_38375574
|
0.007
|
ENST00000323571.4
ENST00000585043.1
ENST00000394103.3
ENST00000536600.1
|
WIPF2
|
WAS/WASL interacting protein family, member 2
|
|
chr2_+_189839046
|
0.007
|
ENST00000304636.3
ENST00000317840.5
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_204801471
|
0.007
|
ENST00000316386.6
ENST00000435193.1
|
ICOS
|
inducible T-cell co-stimulator
|
|
chr9_-_135819987
|
0.004
|
ENST00000298552.3
ENST00000403810.1
|
TSC1
|
tuberous sclerosis 1
|
|
chr15_-_65715401
|
0.004
|
ENST00000352385.2
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4
|
|
chr12_-_57472522
|
0.004
|
ENST00000379391.3
ENST00000300128.4
|
TMEM194A
|
transmembrane protein 194A
|
|
chr9_+_99212403
|
0.004
|
ENST00000375251.3
ENST00000375249.4
|
HABP4
|
hyaluronan binding protein 4
|
|
chr1_+_164528866
|
0.004
|
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr9_-_72287191
|
0.004
|
ENST00000265381.4
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr12_-_42877764
|
0.004
|
ENST00000455697.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr7_+_30067973
|
0.003
|
ENST00000258679.7
ENST00000449726.1
ENST00000396257.2
ENST00000396259.1
|
PLEKHA8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8
|
|
chr22_+_30279144
|
0.003
|
ENST00000401950.2
ENST00000333027.3
ENST00000445401.1
ENST00000323630.5
ENST00000351488.3
|
MTMR3
|
myotubularin related protein 3
|
|
chr12_-_58240470
|
0.003
|
ENST00000548823.1
ENST00000398073.2
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2
|
|
chr2_+_160568978
|
0.003
|
ENST00000409175.1
ENST00000539065.1
ENST00000259050.4
ENST00000421037.1
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase
|
|
chr15_+_45879321
|
0.002
|
ENST00000220531.3
ENST00000567461.1
|
BLOC1S6
|
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin
|
|
chr14_+_55493920
|
0.002
|
ENST00000395472.2
ENST00000555846.1
|
SOCS4
|
suppressor of cytokine signaling 4
|
|
chr10_+_72238517
|
0.002
|
ENST00000263563.6
|
PALD1
|
phosphatase domain containing, paladin 1
|
|
chr8_+_121137333
|
0.002
|
ENST00000309791.4
ENST00000297848.3
ENST00000247781.3
|
COL14A1
|
collagen, type XIV, alpha 1
|
|
chr8_+_59465728
|
0.001
|
ENST00000260130.4
ENST00000422546.2
ENST00000447182.2
ENST00000413219.2
ENST00000424270.2
ENST00000523483.1
ENST00000520168.1
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr12_+_62654119
|
0.001
|
ENST00000353364.3
ENST00000549523.1
ENST00000280377.5
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr4_+_55095264
|
0.001
|
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr12_+_12764773
|
0.001
|
ENST00000228865.2
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr1_-_205601064
|
0.001
|
ENST00000357992.4
ENST00000289703.4
|
ELK4
|
ELK4, ETS-domain protein (SRF accessory protein 1)
|
|
chr11_-_132813566
|
0.001
|
ENST00000331898.7
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|