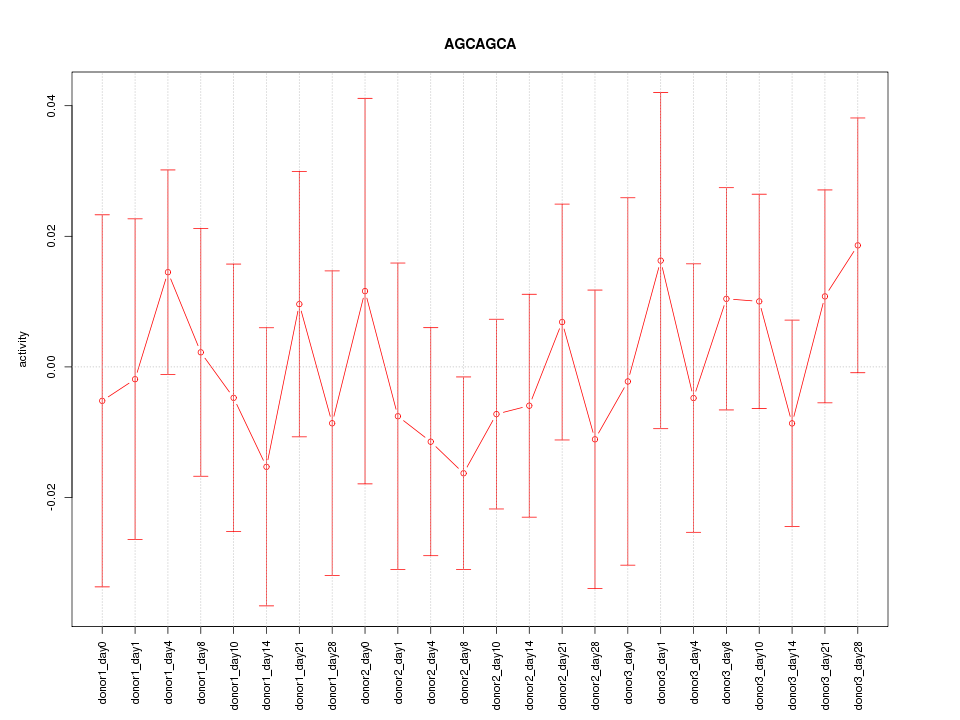
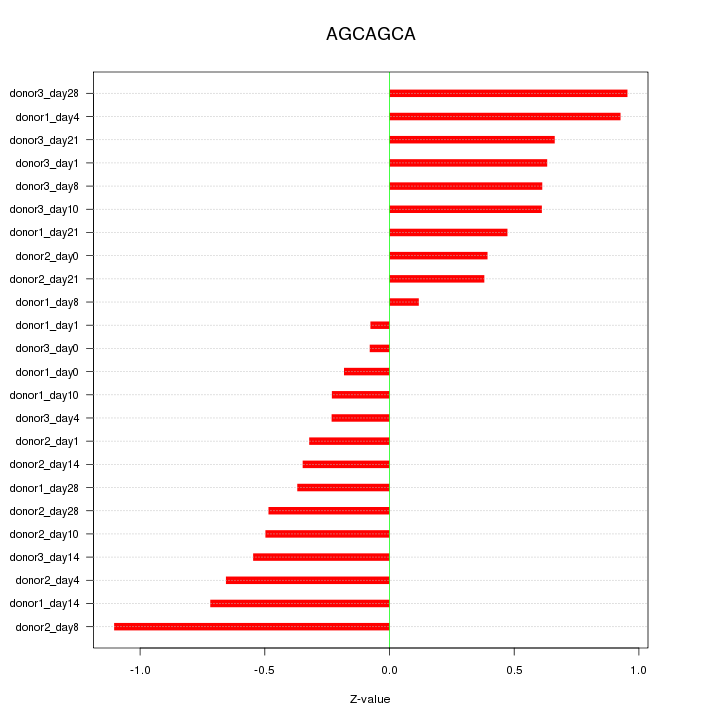
| 0.1 |
0.4 |
GO:0002276 |
basophil activation involved in immune response(GO:0002276) |
| 0.1 |
0.3 |
GO:2001037 |
tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.1 |
0.2 |
GO:0031133 |
regulation of axon diameter(GO:0031133) |
| 0.1 |
0.2 |
GO:1903570 |
regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 |
0.7 |
GO:0016191 |
synaptic vesicle uncoating(GO:0016191) |
| 0.1 |
0.2 |
GO:0021718 |
superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 |
0.3 |
GO:0090402 |
oncogene-induced cell senescence(GO:0090402) |
| 0.1 |
0.3 |
GO:0061767 |
negative regulation of lung blood pressure(GO:0061767) |
| 0.1 |
0.2 |
GO:0032911 |
negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 |
0.2 |
GO:1904647 |
response to rotenone(GO:1904647) |
| 0.1 |
0.2 |
GO:1902534 |
single-organism membrane invagination(GO:1902534) |
| 0.1 |
0.1 |
GO:1903526 |
negative regulation of membrane tubulation(GO:1903526) |
| 0.0 |
0.4 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.2 |
GO:0031064 |
negative regulation of histone deacetylation(GO:0031064) |
| 0.0 |
0.2 |
GO:0051138 |
positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 |
0.1 |
GO:0021503 |
neural fold bending(GO:0021503) |
| 0.0 |
0.1 |
GO:0044805 |
late nucleophagy(GO:0044805) |
| 0.0 |
0.1 |
GO:0098937 |
dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.0 |
0.6 |
GO:0007028 |
cytoplasm organization(GO:0007028) |
| 0.0 |
0.1 |
GO:0071109 |
superior temporal gyrus development(GO:0071109) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 |
0.1 |
GO:1904170 |
regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.0 |
0.2 |
GO:0031585 |
regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) secretory granule localization(GO:0032252) endoplasmic reticulum localization(GO:0051643) |
| 0.0 |
0.1 |
GO:0014057 |
positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 |
0.8 |
GO:0071481 |
cellular response to X-ray(GO:0071481) |
| 0.0 |
0.1 |
GO:1904562 |
phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 |
0.1 |
GO:0002644 |
negative regulation of tolerance induction(GO:0002644) |
| 0.0 |
0.1 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 |
0.1 |
GO:0097051 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 |
0.1 |
GO:2000393 |
negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 |
0.1 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 |
0.4 |
GO:0015939 |
pantothenate metabolic process(GO:0015939) |
| 0.0 |
0.2 |
GO:0051012 |
microtubule sliding(GO:0051012) |
| 0.0 |
0.1 |
GO:2001301 |
lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 |
0.1 |
GO:1903519 |
apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 |
0.1 |
GO:1904504 |
regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 |
0.2 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.0 |
0.1 |
GO:0044837 |
assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 |
0.2 |
GO:0071651 |
positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 |
0.1 |
GO:0032792 |
negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 |
0.1 |
GO:0070541 |
response to platinum ion(GO:0070541) |
| 0.0 |
0.2 |
GO:0002322 |
B cell proliferation involved in immune response(GO:0002322) |
| 0.0 |
0.1 |
GO:1900248 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 |
0.2 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.0 |
0.2 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.0 |
0.2 |
GO:1904627 |
response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 |
0.1 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 |
0.4 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 |
0.2 |
GO:0033625 |
positive regulation of integrin activation(GO:0033625) |
| 0.0 |
0.4 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.0 |
0.1 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.0 |
0.2 |
GO:0070561 |
vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 |
0.1 |
GO:0043397 |
corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 |
0.1 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 |
0.2 |
GO:0060011 |
Sertoli cell proliferation(GO:0060011) |
| 0.0 |
0.1 |
GO:0001555 |
oocyte growth(GO:0001555) |
| 0.0 |
0.2 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.2 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 |
0.1 |
GO:0090170 |
regulation of Golgi inheritance(GO:0090170) |
| 0.0 |
0.2 |
GO:0060591 |
chondroblast differentiation(GO:0060591) |
| 0.0 |
0.1 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 |
0.1 |
GO:1903070 |
negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 |
0.0 |
GO:0097491 |
sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) |
| 0.0 |
0.2 |
GO:0070236 |
negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 |
0.1 |
GO:1903347 |
negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 |
0.1 |
GO:0023021 |
termination of signal transduction(GO:0023021) |
| 0.0 |
0.3 |
GO:0021785 |
branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 |
0.1 |
GO:0035087 |
siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 |
0.1 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) translation reinitiation(GO:0002188) |
| 0.0 |
0.1 |
GO:2001045 |
closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 |
0.2 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.0 |
0.1 |
GO:0030805 |
regulation of cyclic nucleotide catabolic process(GO:0030805) regulation of cAMP catabolic process(GO:0030820) regulation of purine nucleotide catabolic process(GO:0033121) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 |
0.2 |
GO:0070257 |
positive regulation of mucus secretion(GO:0070257) |
| 0.0 |
0.1 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 |
0.0 |
GO:1904760 |
myofibroblast differentiation(GO:0036446) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 |
0.1 |
GO:1990167 |
protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 |
0.0 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 |
0.1 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 |
0.0 |
GO:0035897 |
proteolysis in other organism(GO:0035897) |
| 0.0 |
0.0 |
GO:0090526 |
regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 |
0.1 |
GO:1904075 |
trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 |
0.1 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.0 |
0.3 |
GO:0006751 |
glutathione catabolic process(GO:0006751) |
| 0.0 |
0.1 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.0 |
0.1 |
GO:0032417 |
positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 |
0.0 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 |
0.2 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.0 |
0.0 |
GO:0061534 |
gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 |
0.2 |
GO:0010666 |
positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 |
0.0 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.0 |
0.1 |
GO:0052405 |
negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 |
0.1 |
GO:0042816 |
vitamin B6 metabolic process(GO:0042816) |
| 0.0 |
0.1 |
GO:0007538 |
primary sex determination(GO:0007538) |
| 0.0 |
0.0 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.0 |
0.2 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 |
0.1 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 |
0.2 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.0 |
0.1 |
GO:0030950 |
establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 |
0.0 |
GO:2000276 |
negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 |
0.1 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.0 |
0.0 |
GO:0060067 |
cervix development(GO:0060067) |
| 0.0 |
0.2 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.0 |
GO:0046671 |
negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 |
0.1 |
GO:2000381 |
negative regulation of mesoderm development(GO:2000381) |
| 0.0 |
0.1 |
GO:0035407 |
histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 |
0.1 |
GO:0061087 |
positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 |
0.0 |
GO:0003091 |
renal water homeostasis(GO:0003091) |
| 0.0 |
0.1 |
GO:0061734 |
parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 |
0.1 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 |
0.1 |
GO:1903895 |
negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.0 |
0.1 |
GO:0008204 |
ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 |
0.1 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 |
0.1 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.0 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.0 |
0.2 |
GO:0046541 |
saliva secretion(GO:0046541) |
| 0.0 |
0.1 |
GO:0010868 |
negative regulation of triglyceride biosynthetic process(GO:0010868) |