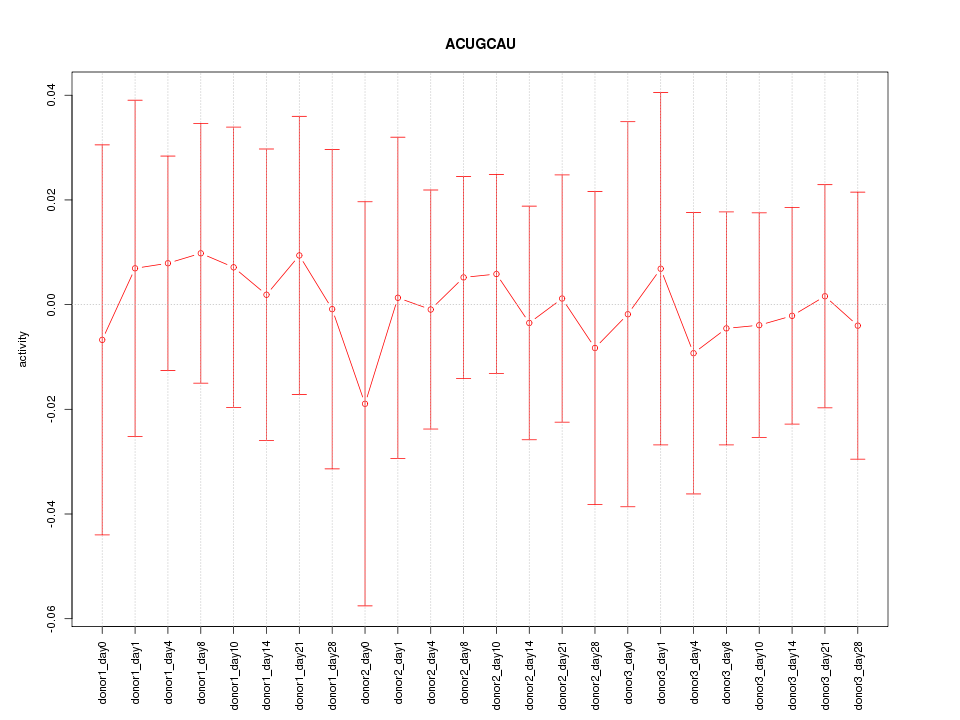
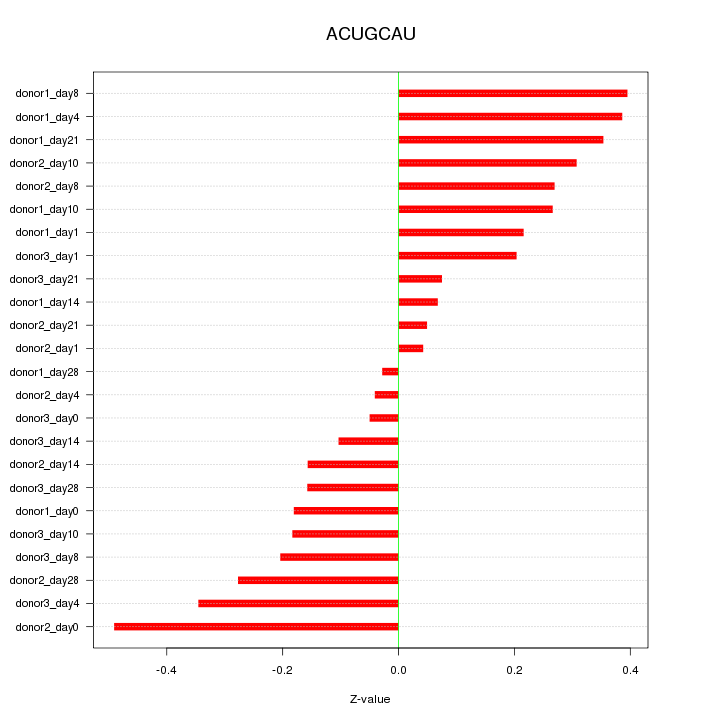
|
chr2_+_39893043
|
0.165
|
ENST00000281961.2
|
TMEM178A
|
transmembrane protein 178A
|
|
chr7_-_122526799
|
0.165
|
ENST00000334010.7
ENST00000313070.7
|
CADPS2
|
Ca++-dependent secretion activator 2
|
|
chr7_-_111846435
|
0.160
|
ENST00000437633.1
ENST00000428084.1
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr17_+_72733350
|
0.157
|
ENST00000392613.5
ENST00000392612.3
ENST00000392610.1
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr7_+_36429409
|
0.127
|
ENST00000265748.2
|
ANLN
|
anillin, actin binding protein
|
|
chr4_-_23891693
|
0.122
|
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr13_+_27131887
|
0.121
|
ENST00000335327.5
|
WASF3
|
WAS protein family, member 3
|
|
chr1_-_217262969
|
0.119
|
ENST00000361525.3
|
ESRRG
|
estrogen-related receptor gamma
|
|
chrX_-_129402857
|
0.113
|
ENST00000447817.1
ENST00000370978.4
|
ZNF280C
|
zinc finger protein 280C
|
|
chr9_-_127703333
|
0.111
|
ENST00000373555.4
|
GOLGA1
|
golgin A1
|
|
chr3_+_155588300
|
0.100
|
ENST00000496455.2
|
GMPS
|
guanine monphosphate synthase
|
|
chr4_+_72204755
|
0.093
|
ENST00000512686.1
ENST00000340595.3
|
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4
|
|
chr15_-_23932437
|
0.092
|
ENST00000331837.4
|
NDN
|
necdin, melanoma antigen (MAGE) family member
|
|
chr20_-_4982132
|
0.088
|
ENST00000338244.1
ENST00000424750.2
|
SLC23A2
|
solute carrier family 23 (ascorbic acid transporter), member 2
|
|
chr17_+_58677539
|
0.086
|
ENST00000305921.3
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr3_-_167813672
|
0.085
|
ENST00000470487.1
|
GOLIM4
|
golgi integral membrane protein 4
|
|
chr5_-_137911049
|
0.085
|
ENST00000297185.3
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin)
|
|
chr10_+_63422695
|
0.084
|
ENST00000330194.2
ENST00000389639.3
|
C10orf107
|
chromosome 10 open reading frame 107
|
|
chr1_-_225840747
|
0.083
|
ENST00000366843.2
ENST00000366844.3
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr6_-_52926539
|
0.083
|
ENST00000350082.5
ENST00000356971.3
|
ICK
|
intestinal cell (MAK-like) kinase
|
|
chr8_-_141645645
|
0.078
|
ENST00000519980.1
ENST00000220592.5
|
AGO2
|
argonaute RISC catalytic component 2
|
|
chr5_-_133304473
|
0.075
|
ENST00000231512.3
|
C5orf15
|
chromosome 5 open reading frame 15
|
|
chr9_+_33025209
|
0.073
|
ENST00000330899.4
ENST00000544625.1
|
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chr6_+_69345166
|
0.072
|
ENST00000370598.1
|
BAI3
|
brain-specific angiogenesis inhibitor 3
|
|
chr9_-_15510989
|
0.070
|
ENST00000380715.1
ENST00000380716.4
ENST00000380738.4
ENST00000380733.4
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr15_-_101792137
|
0.068
|
ENST00000254190.3
|
CHSY1
|
chondroitin sulfate synthase 1
|
|
chr2_-_24149977
|
0.064
|
ENST00000238789.5
|
ATAD2B
|
ATPase family, AAA domain containing 2B
|
|
chr13_+_39261224
|
0.063
|
ENST00000280481.7
|
FREM2
|
FRAS1 related extracellular matrix protein 2
|
|
chr20_+_10199468
|
0.061
|
ENST00000254976.2
ENST00000304886.2
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr1_+_214161272
|
0.059
|
ENST00000498508.2
ENST00000366958.4
|
PROX1
|
prospero homeobox 1
|
|
chr16_-_53737795
|
0.057
|
ENST00000262135.4
ENST00000564374.1
ENST00000566096.1
|
RPGRIP1L
|
RPGRIP1-like
|
|
chr3_-_120461378
|
0.054
|
ENST00000273375.3
|
RABL3
|
RAB, member of RAS oncogene family-like 3
|
|
chr1_-_169455169
|
0.054
|
ENST00000367804.4
ENST00000236137.5
|
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2
|
|
chr15_+_57210818
|
0.053
|
ENST00000438423.2
ENST00000267811.5
ENST00000452095.2
ENST00000559609.1
ENST00000333725.5
|
TCF12
|
transcription factor 12
|
|
chr2_-_157189180
|
0.047
|
ENST00000539077.1
ENST00000424077.1
ENST00000426264.1
ENST00000339562.4
ENST00000421709.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr9_-_86432547
|
0.047
|
ENST00000376365.3
ENST00000376371.2
|
GKAP1
|
G kinase anchoring protein 1
|
|
chr17_+_60536002
|
0.045
|
ENST00000582809.1
|
TLK2
|
tousled-like kinase 2
|
|
chr10_-_32636106
|
0.045
|
ENST00000263062.8
ENST00000319778.6
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr2_+_139259324
|
0.042
|
ENST00000280098.4
|
SPOPL
|
speckle-type POZ protein-like
|
|
chr18_-_74207146
|
0.040
|
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516
|
|
chr16_+_77822427
|
0.039
|
ENST00000302536.2
|
VAT1L
|
vesicle amine transport 1-like
|
|
chr2_+_228336849
|
0.038
|
ENST00000409979.2
ENST00000310078.8
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr1_+_93544791
|
0.038
|
ENST00000545708.1
ENST00000540243.1
ENST00000370298.4
|
MTF2
|
metal response element binding transcription factor 2
|
|
chr17_-_60005365
|
0.037
|
ENST00000444766.3
|
INTS2
|
integrator complex subunit 2
|
|
chr2_+_109335929
|
0.037
|
ENST00000283195.6
|
RANBP2
|
RAN binding protein 2
|
|
chr7_+_30174426
|
0.035
|
ENST00000324453.8
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus)
|
|
chr2_+_235860616
|
0.035
|
ENST00000392011.2
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr3_+_40351169
|
0.035
|
ENST00000232905.3
|
EIF1B
|
eukaryotic translation initiation factor 1B
|
|
chr13_-_21476900
|
0.035
|
ENST00000400602.2
ENST00000255305.6
|
XPO4
|
exportin 4
|
|
chr20_-_44991813
|
0.035
|
ENST00000372227.1
|
SLC35C2
|
solute carrier family 35 (GDP-fucose transporter), member C2
|
|
chr15_-_44486632
|
0.032
|
ENST00000484674.1
|
FRMD5
|
FERM domain containing 5
|
|
chr13_-_72441315
|
0.032
|
ENST00000305425.4
ENST00000313174.7
ENST00000354591.4
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr5_+_143584814
|
0.031
|
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16
|
|
chr6_+_126112001
|
0.031
|
ENST00000392477.2
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr21_-_43430440
|
0.031
|
ENST00000398505.3
ENST00000310826.5
ENST00000449949.1
ENST00000398499.1
ENST00000398497.2
ENST00000398511.3
|
ZBTB21
|
zinc finger and BTB domain containing 21
|
|
chr2_+_99953816
|
0.031
|
ENST00000289371.6
|
EIF5B
|
eukaryotic translation initiation factor 5B
|
|
chr4_-_125633876
|
0.031
|
ENST00000504087.1
ENST00000515641.1
|
ANKRD50
|
ankyrin repeat domain 50
|
|
chr9_-_6007787
|
0.030
|
ENST00000399933.3
ENST00000381461.2
ENST00000513355.2
|
KIAA2026
|
KIAA2026
|
|
chr3_-_79068594
|
0.030
|
ENST00000436010.2
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr5_+_112312416
|
0.030
|
ENST00000389063.2
|
DCP2
|
decapping mRNA 2
|
|
chr2_+_191273052
|
0.029
|
ENST00000417958.1
ENST00000432036.1
ENST00000392328.1
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr11_-_67980744
|
0.029
|
ENST00000401547.2
ENST00000453170.1
ENST00000304363.4
|
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila)
|
|
chr16_-_87739290
|
0.028
|
ENST00000446344.1
|
FLJ00104
|
HCG1980662; Uncharacterized protein
|
|
chr10_-_121356007
|
0.028
|
ENST00000369093.2
ENST00000436547.2
|
TIAL1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1
|
|
chr3_-_18466787
|
0.027
|
ENST00000338745.6
ENST00000450898.1
|
SATB1
|
SATB homeobox 1
|
|
chr10_+_69644404
|
0.027
|
ENST00000212015.6
|
SIRT1
|
sirtuin 1
|
|
chr2_-_86564776
|
0.026
|
ENST00000165698.5
ENST00000541910.1
ENST00000535845.1
|
REEP1
|
receptor accessory protein 1
|
|
chrX_-_134049262
|
0.026
|
ENST00000370783.3
|
MOSPD1
|
motile sperm domain containing 1
|
|
chr4_+_184826418
|
0.026
|
ENST00000308497.4
ENST00000438269.1
|
STOX2
|
storkhead box 2
|
|
chr1_-_244615425
|
0.025
|
ENST00000366535.3
|
ADSS
|
adenylosuccinate synthase
|
|
chr3_-_101232019
|
0.025
|
ENST00000394095.2
ENST00000394091.1
ENST00000394094.2
ENST00000358203.3
ENST00000348610.3
ENST00000314261.7
|
SENP7
|
SUMO1/sentrin specific peptidase 7
|
|
chr20_+_43514315
|
0.024
|
ENST00000353703.4
|
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta
|
|
chr1_-_153895377
|
0.024
|
ENST00000368655.4
|
GATAD2B
|
GATA zinc finger domain containing 2B
|
|
chr1_-_70671216
|
0.024
|
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40
|
|
chr10_+_54074033
|
0.024
|
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1
|
|
chr16_+_16043406
|
0.024
|
ENST00000399410.3
ENST00000399408.2
ENST00000346370.5
ENST00000351154.5
ENST00000345148.5
ENST00000349029.5
|
ABCC1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 1
|
|
chr9_+_129677039
|
0.023
|
ENST00000259351.5
ENST00000424082.2
ENST00000394022.3
ENST00000394011.3
ENST00000319107.4
|
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1
|
|
chr12_+_104359576
|
0.023
|
ENST00000392872.3
ENST00000436021.2
|
TDG
|
thymine-DNA glycosylase
|
|
chr2_+_54951679
|
0.022
|
ENST00000356458.6
|
EML6
|
echinoderm microtubule associated protein like 6
|
|
chr19_+_41768401
|
0.021
|
ENST00000352456.3
ENST00000595018.1
ENST00000597725.1
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr2_+_178077477
|
0.021
|
ENST00000411529.2
ENST00000435711.1
|
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3
|
|
chr10_-_105615164
|
0.021
|
ENST00000355946.2
ENST00000369774.4
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr1_-_35658736
|
0.021
|
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr10_+_43633914
|
0.020
|
ENST00000374466.3
ENST00000374464.1
|
CSGALNACT2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2
|
|
chr6_+_119215308
|
0.020
|
ENST00000229595.5
|
ASF1A
|
anti-silencing function 1A histone chaperone
|
|
chrX_-_73834449
|
0.020
|
ENST00000332687.6
ENST00000349225.2
|
RLIM
|
ring finger protein, LIM domain interacting
|
|
chr12_-_76478686
|
0.020
|
ENST00000261182.8
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr1_+_109234907
|
0.020
|
ENST00000370025.4
ENST00000370022.5
ENST00000370021.1
|
PRPF38B
|
pre-mRNA processing factor 38B
|
|
chr6_-_79944336
|
0.019
|
ENST00000344726.5
ENST00000275036.7
|
HMGN3
|
high mobility group nucleosomal binding domain 3
|
|
chr3_-_44519131
|
0.019
|
ENST00000425708.2
ENST00000396077.2
|
ZNF445
|
zinc finger protein 445
|
|
chr13_+_50656307
|
0.018
|
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding)
|
|
chr5_+_134181625
|
0.018
|
ENST00000394976.3
|
C5orf24
|
chromosome 5 open reading frame 24
|
|
chr9_+_140513438
|
0.018
|
ENST00000462484.1
ENST00000334856.6
ENST00000460843.1
|
EHMT1
|
euchromatic histone-lysine N-methyltransferase 1
|
|
chr5_-_78809950
|
0.018
|
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila)
|
|
chr12_-_109125285
|
0.018
|
ENST00000552871.1
ENST00000261401.3
|
CORO1C
|
coronin, actin binding protein, 1C
|
|
chr9_-_126030817
|
0.018
|
ENST00000348403.5
ENST00000447404.2
ENST00000360998.3
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr3_-_114790179
|
0.017
|
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr8_-_103876965
|
0.017
|
ENST00000337198.5
|
AZIN1
|
antizyme inhibitor 1
|
|
chr8_-_74884511
|
0.017
|
ENST00000518127.1
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C)
|
|
chr2_-_100721178
|
0.017
|
ENST00000409236.2
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chrX_-_135056216
|
0.017
|
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1
|
|
chr13_-_30424821
|
0.016
|
ENST00000380680.4
|
UBL3
|
ubiquitin-like 3
|
|
chr10_+_11206925
|
0.016
|
ENST00000354440.2
ENST00000315874.4
ENST00000427450.1
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr6_+_64281906
|
0.016
|
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr9_+_108210279
|
0.015
|
ENST00000374716.4
ENST00000374710.3
ENST00000481272.1
ENST00000484973.1
ENST00000394926.3
ENST00000539376.1
|
FSD1L
|
fibronectin type III and SPRY domain containing 1-like
|
|
chr10_-_98346801
|
0.014
|
ENST00000371142.4
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr1_-_24306798
|
0.014
|
ENST00000374452.5
ENST00000492112.2
ENST00000343255.5
ENST00000344989.6
|
SRSF10
|
serine/arginine-rich splicing factor 10
|
|
chr14_-_24020858
|
0.014
|
ENST00000419474.3
|
ZFHX2
|
zinc finger homeobox 2
|
|
chr4_+_57774042
|
0.014
|
ENST00000309042.7
|
REST
|
RE1-silencing transcription factor
|
|
chr15_+_44829255
|
0.013
|
ENST00000261868.5
ENST00000424492.3
|
EIF3J
|
eukaryotic translation initiation factor 3, subunit J
|
|
chr17_+_29421900
|
0.013
|
ENST00000358273.4
ENST00000356175.3
|
NF1
|
neurofibromin 1
|
|
chr17_+_30264014
|
0.013
|
ENST00000322652.5
ENST00000580398.1
|
SUZ12
|
SUZ12 polycomb repressive complex 2 subunit
|
|
chr7_-_75988321
|
0.013
|
ENST00000307630.3
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma
|
|
chr1_-_233431458
|
0.013
|
ENST00000258229.9
ENST00000430153.1
|
PCNXL2
|
pecanex-like 2 (Drosophila)
|
|
chr12_+_72233487
|
0.013
|
ENST00000482439.2
ENST00000550746.1
ENST00000491063.1
ENST00000319106.8
ENST00000485960.2
ENST00000393309.3
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr4_+_144106080
|
0.013
|
ENST00000307017.4
|
USP38
|
ubiquitin specific peptidase 38
|
|
chr2_+_124782857
|
0.012
|
ENST00000431078.1
|
CNTNAP5
|
contactin associated protein-like 5
|
|
chr5_-_130970723
|
0.012
|
ENST00000308008.6
ENST00000296859.6
ENST00000507093.1
ENST00000510071.1
ENST00000509018.1
ENST00000307984.5
|
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6
|
|
chr10_+_114709999
|
0.012
|
ENST00000355995.4
ENST00000545257.1
ENST00000543371.1
ENST00000536810.1
ENST00000355717.4
ENST00000538897.1
ENST00000534894.1
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr3_+_113465866
|
0.011
|
ENST00000273398.3
ENST00000538620.1
ENST00000496747.1
ENST00000475322.1
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr20_+_3451650
|
0.011
|
ENST00000262919.5
|
ATRN
|
attractin
|
|
chr12_+_104850740
|
0.011
|
ENST00000547956.1
ENST00000549260.1
ENST00000303694.5
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr16_-_80838195
|
0.011
|
ENST00000570137.2
|
CDYL2
|
chromodomain protein, Y-like 2
|
|
chr3_+_30648066
|
0.011
|
ENST00000359013.4
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr3_+_180630090
|
0.011
|
ENST00000357559.4
ENST00000305586.7
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr12_+_69004619
|
0.010
|
ENST00000250559.9
ENST00000393436.5
ENST00000425247.2
ENST00000489473.2
ENST00000422358.2
ENST00000541167.1
ENST00000538283.1
ENST00000341355.5
ENST00000537460.1
ENST00000450214.2
ENST00000545270.1
ENST00000538980.1
ENST00000542018.1
ENST00000543393.1
|
RAP1B
|
RAP1B, member of RAS oncogene family
|
|
chr11_+_125462690
|
0.010
|
ENST00000392708.4
ENST00000529196.1
ENST00000531491.1
|
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic)
|
|
chr3_-_48342795
|
0.010
|
ENST00000452211.1
ENST00000447724.1
ENST00000415053.1
ENST00000425930.1
ENST00000421967.1
ENST00000426723.1
ENST00000450160.1
ENST00000456495.1
ENST00000435684.1
ENST00000447314.1
ENST00000415644.1
ENST00000442597.1
|
NME6
|
NME/NM23 nucleoside diphosphate kinase 6
|
|
chr4_+_77870856
|
0.010
|
ENST00000264893.6
ENST00000502584.1
ENST00000510641.1
|
SEPT11
|
septin 11
|
|
chr16_+_56965960
|
0.009
|
ENST00000439977.2
ENST00000344114.4
ENST00000300302.5
ENST00000379792.2
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr13_+_33160553
|
0.009
|
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae)
|
|
chr12_+_123237321
|
0.008
|
ENST00000280557.6
ENST00000455982.2
|
DENR
|
density-regulated protein
|
|
chr19_-_47616992
|
0.008
|
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chrX_-_112084043
|
0.008
|
ENST00000304758.1
|
AMOT
|
angiomotin
|
|
chr5_+_139027877
|
0.008
|
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5
|
|
chr3_+_11034403
|
0.008
|
ENST00000287766.4
ENST00000425938.1
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter), member 1
|
|
chr1_-_179198702
|
0.008
|
ENST00000502732.1
|
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase
|
|
chrX_+_12993202
|
0.007
|
ENST00000451311.2
ENST00000380636.1
|
TMSB4X
|
thymosin beta 4, X-linked
|
|
chr1_+_151584544
|
0.007
|
ENST00000458013.2
ENST00000368843.3
|
SNX27
|
sorting nexin family member 27
|
|
chr10_+_23728198
|
0.007
|
ENST00000376495.3
|
OTUD1
|
OTU domain containing 1
|
|
chr5_-_131132658
|
0.007
|
ENST00000511848.1
ENST00000510461.1
ENST00000514667.1
|
FNIP1
CTC-432M15.3
|
folliculin interacting protein 1
Folliculin-interacting protein 1
|
|
chr13_+_58206655
|
0.007
|
ENST00000377918.3
|
PCDH17
|
protocadherin 17
|
|
chr5_-_56247935
|
0.006
|
ENST00000381199.3
ENST00000381226.3
ENST00000381213.3
|
MIER3
|
mesoderm induction early response 1, family member 3
|
|
chr8_-_29940628
|
0.006
|
ENST00000545648.1
ENST00000256255.6
|
TMEM66
|
transmembrane protein 66
|
|
chr12_-_49449107
|
0.006
|
ENST00000301067.7
|
KMT2D
|
lysine (K)-specific methyltransferase 2D
|
|
chr6_-_166796461
|
0.006
|
ENST00000360961.6
ENST00000341756.6
|
MPC1
|
mitochondrial pyruvate carrier 1
|
|
chr10_-_27443155
|
0.006
|
ENST00000427324.1
ENST00000326799.3
|
YME1L1
|
YME1-like 1 ATPase
|
|
chr6_+_111195973
|
0.006
|
ENST00000368885.3
ENST00000368882.3
ENST00000451850.2
ENST00000368877.5
|
AMD1
|
adenosylmethionine decarboxylase 1
|
|
chr7_-_83824169
|
0.006
|
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr16_+_22019404
|
0.005
|
ENST00000542527.2
ENST00000569656.1
ENST00000562695.1
|
C16orf52
|
chromosome 16 open reading frame 52
|
|
chr5_+_61602055
|
0.005
|
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A
|
|
chr18_+_43753974
|
0.005
|
ENST00000282059.6
ENST00000321319.6
|
C18orf25
|
chromosome 18 open reading frame 25
|
|
chr13_+_42622781
|
0.005
|
ENST00000337343.4
ENST00000261491.5
ENST00000379274.2
|
DGKH
|
diacylglycerol kinase, eta
|
|
chr2_+_169312350
|
0.005
|
ENST00000305747.6
|
CERS6
|
ceramide synthase 6
|
|
chr5_+_36876833
|
0.005
|
ENST00000282516.8
ENST00000448238.2
|
NIPBL
|
Nipped-B homolog (Drosophila)
|
|
chr15_-_83621435
|
0.005
|
ENST00000450735.2
ENST00000426485.1
ENST00000399166.2
ENST00000304231.8
|
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr2_+_196521458
|
0.005
|
ENST00000409086.3
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr8_+_98656336
|
0.004
|
ENST00000336273.3
|
MTDH
|
metadherin
|
|
chr12_+_19592602
|
0.004
|
ENST00000398864.3
ENST00000266508.9
|
AEBP2
|
AE binding protein 2
|
|
chr17_-_60142609
|
0.004
|
ENST00000397786.2
|
MED13
|
mediator complex subunit 13
|
|
chr5_-_168006591
|
0.004
|
ENST00000239231.6
|
PANK3
|
pantothenate kinase 3
|
|
chr15_+_44719394
|
0.003
|
ENST00000260327.4
ENST00000396780.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2
|
|
chr2_+_26568965
|
0.003
|
ENST00000260585.7
ENST00000447170.1
|
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific)
|
|
chr3_-_53925863
|
0.003
|
ENST00000541726.1
ENST00000495461.1
|
SELK
|
Selenoprotein K
|
|
chr17_+_41052808
|
0.003
|
ENST00000592383.1
ENST00000253801.2
ENST00000585489.1
|
G6PC
|
glucose-6-phosphatase, catalytic subunit
|
|
chrX_+_9983602
|
0.003
|
ENST00000380861.4
|
WWC3
|
WWC family member 3
|
|
chr12_-_25403737
|
0.003
|
ENST00000256078.4
ENST00000556131.1
ENST00000311936.3
ENST00000557334.1
|
KRAS
|
Kirsten rat sarcoma viral oncogene homolog
|
|
chr20_-_50384864
|
0.003
|
ENST00000311637.5
ENST00000402822.1
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr17_+_38219063
|
0.003
|
ENST00000584985.1
ENST00000264637.4
ENST00000450525.2
|
THRA
|
thyroid hormone receptor, alpha
|
|
chr17_-_58603568
|
0.003
|
ENST00000083182.3
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr9_-_95055956
|
0.002
|
ENST00000375629.3
ENST00000447699.2
ENST00000375643.3
ENST00000395554.3
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr12_-_42538657
|
0.002
|
ENST00000398675.3
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr12_-_42632016
|
0.002
|
ENST00000442791.3
ENST00000327791.4
ENST00000534854.2
ENST00000380788.3
ENST00000380790.4
|
YAF2
|
YY1 associated factor 2
|
|
chr20_+_1099233
|
0.002
|
ENST00000246015.4
ENST00000335877.6
ENST00000438768.2
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31)
|
|
chr9_-_104249319
|
0.002
|
ENST00000374847.1
|
TMEM246
|
transmembrane protein 246
|
|
chr4_-_66536057
|
0.001
|
ENST00000273854.3
|
EPHA5
|
EPH receptor A5
|
|
chr12_-_31479045
|
0.000
|
ENST00000539409.1
ENST00000395766.1
|
FAM60A
|
family with sequence similarity 60, member A
|