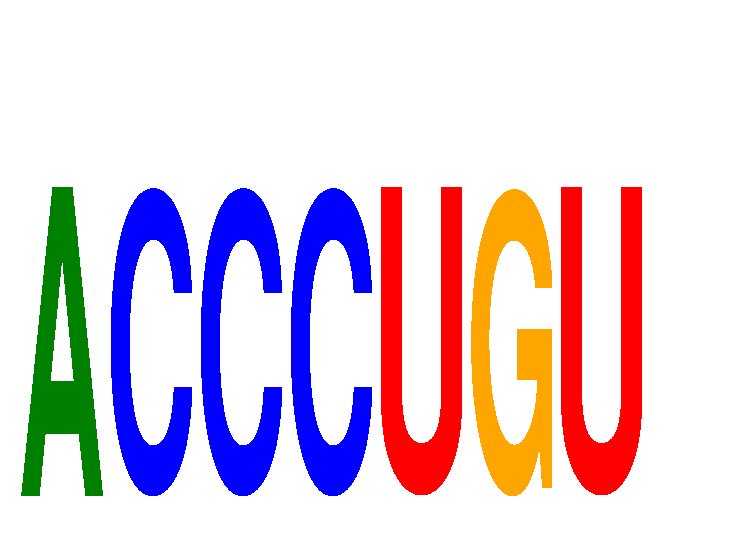|
chr15_-_61521495
|
0.264
|
ENST00000335670.6
|
RORA
|
RAR-related orphan receptor A
|
|
chr3_+_238273
|
0.197
|
ENST00000256509.2
|
CHL1
|
cell adhesion molecule L1-like
|
|
chr1_-_217262969
|
0.189
|
ENST00000361525.3
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr1_+_210406121
|
0.151
|
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4
|
|
chr5_-_124080203
|
0.143
|
ENST00000504926.1
|
ZNF608
|
zinc finger protein 608
|
|
chr3_-_9994021
|
0.133
|
ENST00000411976.2
ENST00000412055.1
|
PRRT3
|
proline-rich transmembrane protein 3
|
|
chr6_+_107811162
|
0.129
|
ENST00000317357.5
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr2_+_10183651
|
0.127
|
ENST00000305883.1
|
KLF11
|
Kruppel-like factor 11
|
|
chr10_-_62149433
|
0.122
|
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr2_+_121103706
|
0.116
|
ENST00000295228.3
|
INHBB
|
inhibin, beta B
|
|
chr19_+_10527449
|
0.112
|
ENST00000592685.1
ENST00000380702.2
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr19_+_13135386
|
0.104
|
ENST00000360105.4
ENST00000588228.1
ENST00000591028.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr7_+_94139105
|
0.103
|
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1
|
|
chr1_+_110082487
|
0.088
|
ENST00000527748.1
|
GPR61
|
G protein-coupled receptor 61
|
|
chr16_+_57126428
|
0.083
|
ENST00000290776.8
|
CPNE2
|
copine II
|
|
chr6_+_117002339
|
0.083
|
ENST00000413340.1
ENST00000368564.1
ENST00000356348.1
|
KPNA5
|
karyopherin alpha 5 (importin alpha 6)
|
|
chr16_-_70719925
|
0.080
|
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr11_-_115375107
|
0.076
|
ENST00000545380.1
ENST00000452722.3
ENST00000537058.1
ENST00000536727.1
ENST00000542447.2
ENST00000331581.6
|
CADM1
|
cell adhesion molecule 1
|
|
chr1_-_39339777
|
0.074
|
ENST00000397572.2
|
MYCBP
|
MYC binding protein
|
|
chr16_+_69599861
|
0.074
|
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr1_-_94374946
|
0.072
|
ENST00000370238.3
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr1_+_28052456
|
0.071
|
ENST00000373954.6
ENST00000419687.2
|
FAM76A
|
family with sequence similarity 76, member A
|
|
chr1_+_57110972
|
0.071
|
ENST00000371244.4
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit
|
|
chr22_-_17602200
|
0.070
|
ENST00000399875.1
|
CECR6
|
cat eye syndrome chromosome region, candidate 6
|
|
chr5_-_160973649
|
0.068
|
ENST00000393959.1
ENST00000517547.1
|
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2
|
|
chr4_-_100575781
|
0.067
|
ENST00000511828.1
|
RP11-766F14.2
|
Protein LOC285556
|
|
chr6_+_116421976
|
0.064
|
ENST00000319550.4
ENST00000419791.1
|
NT5DC1
|
5'-nucleotidase domain containing 1
|
|
chr14_+_45366472
|
0.059
|
ENST00000325192.3
|
C14orf28
|
chromosome 14 open reading frame 28
|
|
chr6_-_91006461
|
0.058
|
ENST00000257749.4
ENST00000343122.3
ENST00000406998.2
ENST00000453877.1
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr18_+_9136758
|
0.058
|
ENST00000383440.2
ENST00000262126.4
ENST00000577992.1
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr9_-_127533519
|
0.056
|
ENST00000487099.2
ENST00000344523.4
ENST00000373584.3
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chrX_-_129402857
|
0.054
|
ENST00000447817.1
ENST00000370978.4
|
ZNF280C
|
zinc finger protein 280C
|
|
chr4_-_53525406
|
0.052
|
ENST00000451218.2
ENST00000441222.3
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr19_-_55954230
|
0.052
|
ENST00000376325.4
|
SHISA7
|
shisa family member 7
|
|
chr5_+_56205081
|
0.049
|
ENST00000285947.2
ENST00000541720.1
|
SETD9
|
SET domain containing 9
|
|
chr7_+_20370746
|
0.048
|
ENST00000222573.4
|
ITGB8
|
integrin, beta 8
|
|
chr16_+_50187556
|
0.047
|
ENST00000561678.1
ENST00000357464.3
|
PAPD5
|
PAP associated domain containing 5
|
|
chr11_+_18344106
|
0.044
|
ENST00000534641.1
ENST00000525831.1
ENST00000265963.4
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr1_+_93913713
|
0.044
|
ENST00000604705.1
ENST00000370253.2
|
FNBP1L
|
formin binding protein 1-like
|
|
chr5_-_45696253
|
0.043
|
ENST00000303230.4
|
HCN1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1
|
|
chr10_+_180987
|
0.042
|
ENST00000381591.1
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr2_-_160472952
|
0.041
|
ENST00000541068.2
ENST00000355831.2
ENST00000343439.5
ENST00000392782.1
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr13_-_29069232
|
0.038
|
ENST00000282397.4
ENST00000541932.1
ENST00000539099.1
|
FLT1
|
fms-related tyrosine kinase 1
|
|
chr7_-_44924939
|
0.038
|
ENST00000395699.2
|
PURB
|
purine-rich element binding protein B
|
|
chr2_+_208394616
|
0.038
|
ENST00000432329.2
ENST00000353267.3
ENST00000445803.1
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr12_+_69864129
|
0.036
|
ENST00000547219.1
ENST00000299293.2
ENST00000549921.1
ENST00000550316.1
ENST00000548154.1
ENST00000547414.1
ENST00000550389.1
ENST00000550937.1
ENST00000549092.1
ENST00000550169.1
|
FRS2
|
fibroblast growth factor receptor substrate 2
|
|
chr1_-_19283163
|
0.036
|
ENST00000455833.2
|
IFFO2
|
intermediate filament family orphan 2
|
|
chr3_+_14989076
|
0.035
|
ENST00000413118.1
ENST00000425241.1
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2
|
|
chr17_+_45727204
|
0.035
|
ENST00000290158.4
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr3_-_142166904
|
0.034
|
ENST00000264951.4
|
XRN1
|
5'-3' exoribonuclease 1
|
|
chr8_+_1449532
|
0.033
|
ENST00000421627.2
|
DLGAP2
|
discs, large (Drosophila) homolog-associated protein 2
|
|
chrX_+_9431324
|
0.033
|
ENST00000407597.2
ENST00000424279.1
ENST00000536365.1
ENST00000441088.1
ENST00000380961.1
ENST00000415293.1
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr19_-_14316980
|
0.033
|
ENST00000361434.3
ENST00000340736.6
|
LPHN1
|
latrophilin 1
|
|
chr5_-_90679145
|
0.033
|
ENST00000265138.3
|
ARRDC3
|
arrestin domain containing 3
|
|
chr17_+_4487294
|
0.032
|
ENST00000338859.4
|
SMTNL2
|
smoothelin-like 2
|
|
chr10_-_75173785
|
0.032
|
ENST00000535178.1
ENST00000372921.5
ENST00000372919.4
|
ANXA7
|
annexin A7
|
|
chr17_-_48207157
|
0.029
|
ENST00000330175.4
ENST00000503131.1
|
SAMD14
|
sterile alpha motif domain containing 14
|
|
chr20_+_55204351
|
0.027
|
ENST00000201031.2
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma)
|
|
chr2_+_148778570
|
0.027
|
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5
|
|
chr7_-_138666053
|
0.026
|
ENST00000440172.1
ENST00000422774.1
|
KIAA1549
|
KIAA1549
|
|
chr18_-_45456930
|
0.026
|
ENST00000262160.6
ENST00000587269.1
|
SMAD2
|
SMAD family member 2
|
|
chr9_-_130661916
|
0.025
|
ENST00000373142.1
ENST00000373146.1
ENST00000373144.3
|
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6
|
|
chr2_-_206950781
|
0.025
|
ENST00000403263.1
|
INO80D
|
INO80 complex subunit D
|
|
chr5_+_63461642
|
0.024
|
ENST00000296615.6
ENST00000381081.2
ENST00000389100.4
|
RNF180
|
ring finger protein 180
|
|
chr10_-_79686284
|
0.023
|
ENST00000372391.2
ENST00000372388.2
|
DLG5
|
discs, large homolog 5 (Drosophila)
|
|
chr4_-_66536057
|
0.023
|
ENST00000273854.3
|
EPHA5
|
EPH receptor A5
|
|
chr1_-_39347255
|
0.023
|
ENST00000454994.2
ENST00000357771.3
|
GJA9
|
gap junction protein, alpha 9, 59kDa
|
|
chr4_+_154125565
|
0.023
|
ENST00000338700.5
|
TRIM2
|
tripartite motif containing 2
|
|
chr1_+_109632425
|
0.022
|
ENST00000338272.8
|
TMEM167B
|
transmembrane protein 167B
|
|
chr17_-_27621125
|
0.021
|
ENST00000579665.1
ENST00000225388.4
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr22_+_40573921
|
0.017
|
ENST00000454349.2
ENST00000335727.9
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr14_+_23775971
|
0.017
|
ENST00000250405.5
|
BCL2L2
|
BCL2-like 2
|
|
chr1_+_89990431
|
0.016
|
ENST00000330947.2
ENST00000358200.4
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr7_-_112430647
|
0.016
|
ENST00000312814.6
|
TMEM168
|
transmembrane protein 168
|
|
chr11_-_18656028
|
0.016
|
ENST00000336349.5
|
SPTY2D1
|
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae)
|
|
chr2_-_37193606
|
0.015
|
ENST00000379213.2
ENST00000263918.4
|
STRN
|
striatin, calmodulin binding protein
|
|
chr1_+_22889953
|
0.014
|
ENST00000374644.4
ENST00000166244.3
ENST00000538803.1
|
EPHA8
|
EPH receptor A8
|
|
chr6_-_11044509
|
0.014
|
ENST00000354666.3
|
ELOVL2
|
ELOVL fatty acid elongase 2
|
|
chr7_-_72936531
|
0.012
|
ENST00000339594.4
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B
|
|
chr12_-_133405288
|
0.012
|
ENST00000204726.3
|
GOLGA3
|
golgin A3
|
|
chr17_-_76713100
|
0.012
|
ENST00000585509.1
|
CYTH1
|
cytohesin 1
|
|
chr17_+_46125707
|
0.012
|
ENST00000584137.1
ENST00000362042.3
ENST00000585291.1
ENST00000357480.5
|
NFE2L1
|
nuclear factor, erythroid 2-like 1
|
|
chr4_+_184020398
|
0.011
|
ENST00000403733.3
ENST00000378925.3
|
WWC2
|
WW and C2 domain containing 2
|
|
chr10_+_98592009
|
0.011
|
ENST00000540664.1
ENST00000371103.3
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr5_+_149109825
|
0.011
|
ENST00000360453.4
ENST00000394320.3
ENST00000309241.5
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr17_-_73775839
|
0.011
|
ENST00000592643.1
ENST00000591890.1
ENST00000587171.1
ENST00000254810.4
ENST00000589599.1
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr19_+_3880581
|
0.010
|
ENST00000450849.2
ENST00000301260.6
ENST00000398448.3
|
ATCAY
|
ataxia, cerebellar, Cayman type
|
|
chr12_-_14956396
|
0.009
|
ENST00000535328.1
ENST00000261167.2
|
WBP11
|
WW domain binding protein 11
|
|
chr9_-_115983641
|
0.009
|
ENST00000238256.3
|
FKBP15
|
FK506 binding protein 15, 133kDa
|
|
chr17_+_36861735
|
0.008
|
ENST00000378137.5
ENST00000325718.7
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr4_+_134070439
|
0.008
|
ENST00000264360.5
|
PCDH10
|
protocadherin 10
|
|
chr9_-_86595503
|
0.008
|
ENST00000376281.4
ENST00000376264.2
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr9_+_100263912
|
0.008
|
ENST00000259365.4
|
TMOD1
|
tropomodulin 1
|
|
chr22_+_30279144
|
0.007
|
ENST00000401950.2
ENST00000333027.3
ENST00000445401.1
ENST00000323630.5
ENST00000351488.3
|
MTMR3
|
myotubularin related protein 3
|
|
chr22_+_45559722
|
0.007
|
ENST00000347635.4
ENST00000407019.2
ENST00000424634.1
ENST00000417702.1
ENST00000425733.2
ENST00000430547.1
|
NUP50
|
nucleoporin 50kDa
|
|
chr11_+_65101225
|
0.006
|
ENST00000528416.1
ENST00000415073.2
ENST00000252268.4
|
DPF2
|
D4, zinc and double PHD fingers family 2
|
|
chr2_+_166326157
|
0.006
|
ENST00000421875.1
ENST00000314499.7
ENST00000409664.1
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr10_+_89622870
|
0.006
|
ENST00000371953.3
|
PTEN
|
phosphatase and tensin homolog
|
|
chr5_+_143584814
|
0.006
|
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16
|
|
chr2_+_111878483
|
0.005
|
ENST00000308659.8
ENST00000357757.2
ENST00000393253.2
ENST00000337565.5
ENST00000393256.3
|
BCL2L11
|
BCL2-like 11 (apoptosis facilitator)
|
|
chr1_+_36396677
|
0.005
|
ENST00000373191.4
ENST00000397828.2
|
AGO3
|
argonaute RISC catalytic component 3
|
|
chr3_+_183873098
|
0.004
|
ENST00000313143.3
|
DVL3
|
dishevelled segment polarity protein 3
|
|
chr15_+_44719394
|
0.004
|
ENST00000260327.4
ENST00000396780.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2
|
|
chrX_-_54384425
|
0.003
|
ENST00000375169.3
ENST00000354646.2
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chr1_+_202976493
|
0.003
|
ENST00000367242.3
|
TMEM183A
|
transmembrane protein 183A
|
|
chr6_+_71377473
|
0.002
|
ENST00000370452.3
ENST00000316999.5
ENST00000370455.3
|
SMAP1
|
small ArfGAP 1
|
|
chr10_-_30348439
|
0.002
|
ENST00000375377.1
|
KIAA1462
|
KIAA1462
|
|
chr10_-_121632266
|
0.002
|
ENST00000360003.3
ENST00000369077.3
|
MCMBP
|
minichromosome maintenance complex binding protein
|
|
chr4_+_79697495
|
0.001
|
ENST00000502871.1
ENST00000335016.5
|
BMP2K
|
BMP2 inducible kinase
|
|
chr21_+_45285050
|
0.001
|
ENST00000291572.8
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr14_+_33408449
|
0.001
|
ENST00000346562.2
ENST00000341321.4
ENST00000548645.1
ENST00000356141.4
ENST00000357798.5
|
NPAS3
|
neuronal PAS domain protein 3
|