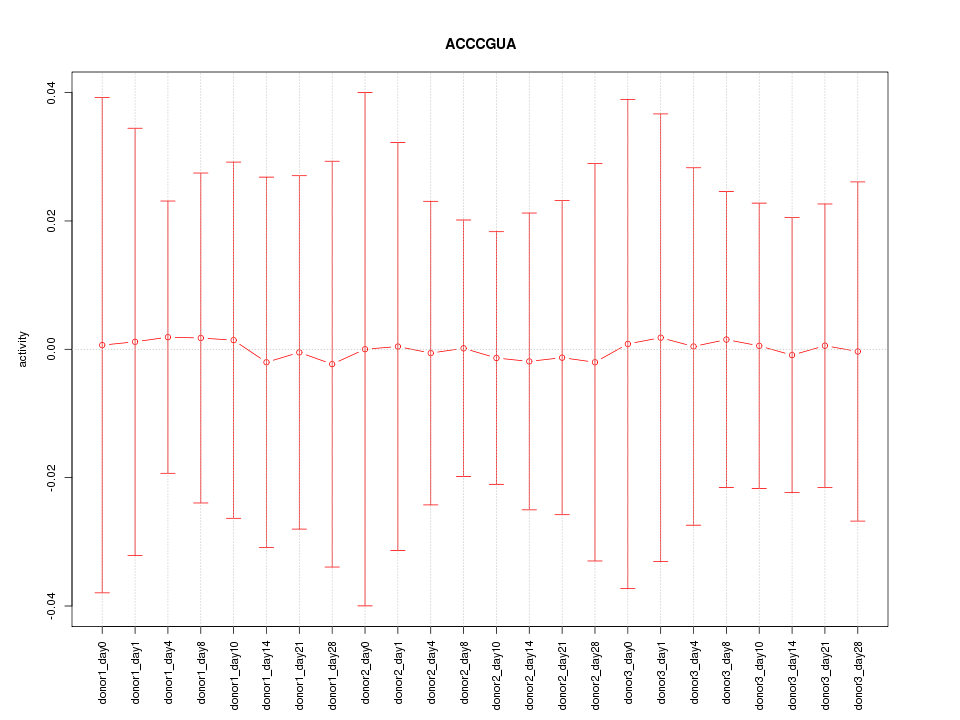
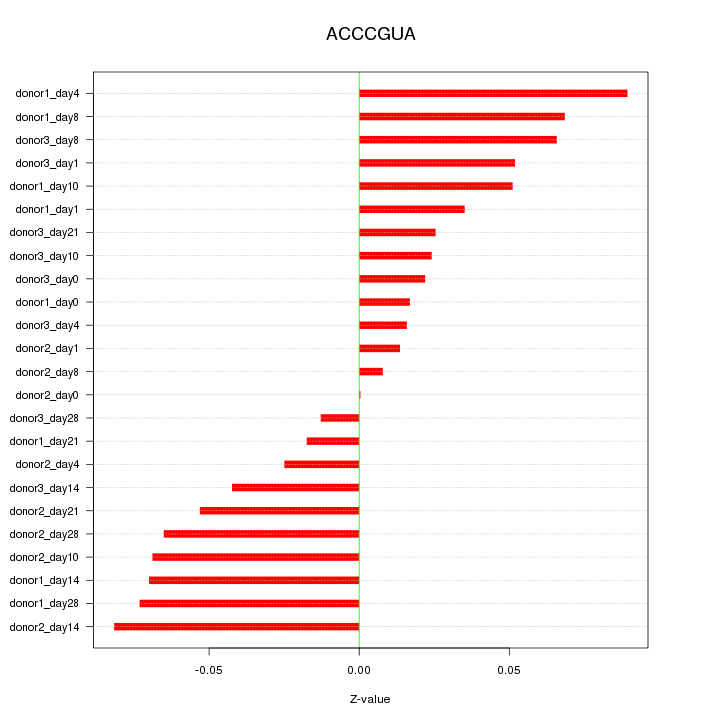
Motif ID: ACCCGUA
Z-value: 0.049
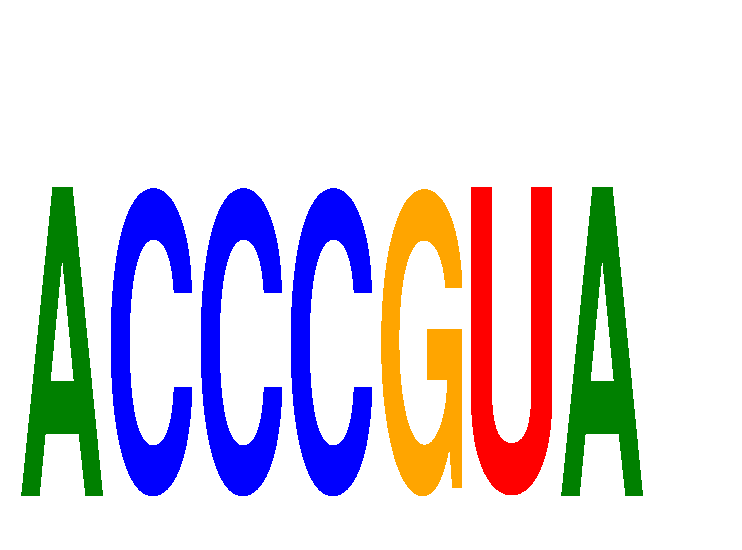
Mature miRNA associated with seed ACCCGUA:
| Name | miRBase Accession |
|---|---|
| hsa-miR-100-5p | MIMAT0000098 |
| hsa-miR-99a-5p | MIMAT0000097 |
| hsa-miR-99b-5p | MIMAT0000689 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |