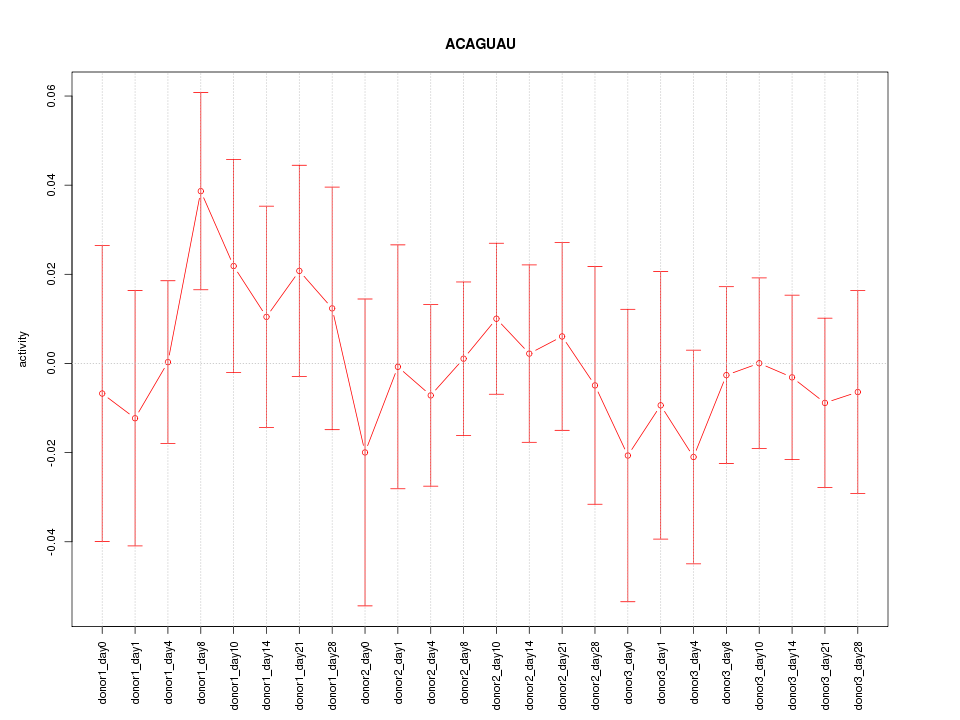
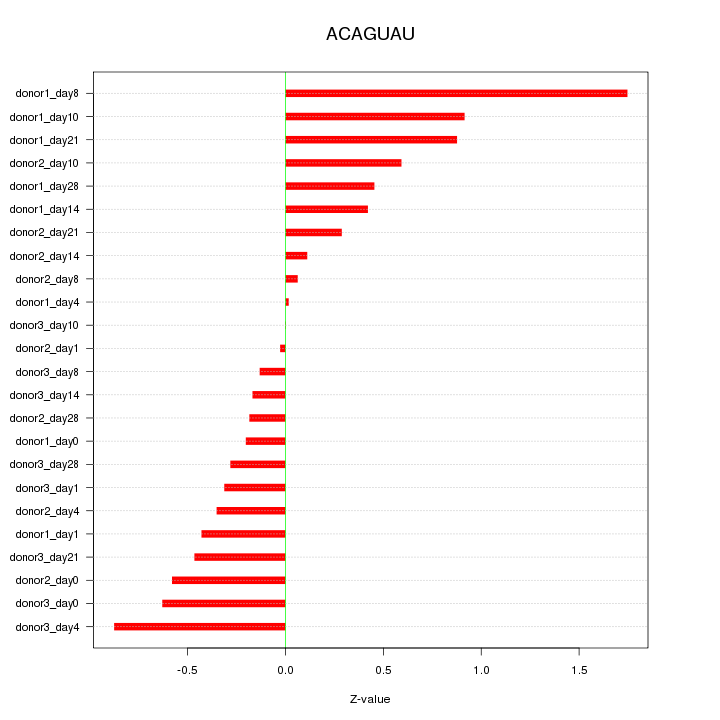
| 0.4 |
1.1 |
GO:0002071 |
glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.2 |
0.7 |
GO:1904899 |
regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.2 |
0.8 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 |
0.9 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 |
0.5 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.2 |
0.5 |
GO:0061181 |
regulation of chondrocyte development(GO:0061181) |
| 0.2 |
0.5 |
GO:1902309 |
negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 |
0.4 |
GO:0003099 |
vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.1 |
0.4 |
GO:2000364 |
regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.1 |
0.6 |
GO:0048861 |
leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 |
0.5 |
GO:0070662 |
mast cell proliferation(GO:0070662) |
| 0.1 |
0.6 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.1 |
0.4 |
GO:0033634 |
positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 |
0.3 |
GO:1902725 |
negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 |
0.3 |
GO:0016260 |
selenocysteine biosynthetic process(GO:0016260) |
| 0.1 |
0.2 |
GO:2000670 |
positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 |
0.3 |
GO:0061428 |
negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 |
0.4 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.1 |
0.4 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 |
0.2 |
GO:1900169 |
regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 |
0.2 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.1 |
0.6 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.1 |
0.2 |
GO:0015993 |
L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 |
0.2 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 |
0.3 |
GO:0035720 |
intraciliary anterograde transport(GO:0035720) |
| 0.1 |
0.2 |
GO:0071879 |
positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 |
0.2 |
GO:0051892 |
negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 |
0.3 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 |
0.4 |
GO:0070086 |
ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 |
0.3 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 |
0.2 |
GO:0035669 |
TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.1 |
0.2 |
GO:0097195 |
pilomotor reflex(GO:0097195) |
| 0.1 |
0.3 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 |
0.1 |
GO:0035936 |
testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.1 |
0.2 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.0 |
0.2 |
GO:0071934 |
thiamine transmembrane transport(GO:0071934) |
| 0.0 |
0.3 |
GO:0060011 |
Sertoli cell proliferation(GO:0060011) |
| 0.0 |
0.2 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 |
0.4 |
GO:0035973 |
aggrephagy(GO:0035973) |
| 0.0 |
0.1 |
GO:1904247 |
positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 |
0.1 |
GO:1901675 |
negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 |
0.1 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.0 |
0.2 |
GO:2000809 |
positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 |
0.3 |
GO:0043634 |
polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.0 |
0.1 |
GO:0061289 |
cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 |
0.2 |
GO:0045629 |
negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 |
0.0 |
GO:1901187 |
regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 |
0.2 |
GO:0001880 |
Mullerian duct regression(GO:0001880) |
| 0.0 |
0.2 |
GO:0071874 |
cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 |
0.2 |
GO:1903788 |
mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 |
0.2 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 |
0.2 |
GO:2000546 |
positive regulation of fat cell proliferation(GO:0070346) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 |
0.3 |
GO:1990253 |
cellular response to leucine starvation(GO:1990253) |
| 0.0 |
0.1 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 |
0.2 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 |
0.1 |
GO:0072658 |
maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 |
0.5 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.0 |
0.1 |
GO:0048294 |
negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 |
0.3 |
GO:1903025 |
regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 |
0.2 |
GO:0086024 |
adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 |
0.1 |
GO:2000645 |
negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 |
0.3 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 |
0.1 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.0 |
0.2 |
GO:1990034 |
cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 |
0.2 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.1 |
GO:1902617 |
response to fluoride(GO:1902617) |
| 0.0 |
0.1 |
GO:0046502 |
uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 |
0.2 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 |
0.1 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 0.0 |
0.3 |
GO:0060536 |
cartilage morphogenesis(GO:0060536) |
| 0.0 |
0.1 |
GO:0033504 |
floor plate development(GO:0033504) |
| 0.0 |
0.3 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.0 |
0.2 |
GO:0061088 |
regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 |
0.5 |
GO:1903204 |
negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 |
0.0 |
GO:1902263 |
apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 |
0.1 |
GO:0035087 |
siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 |
0.1 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.2 |
GO:0036302 |
atrioventricular canal development(GO:0036302) |
| 0.0 |
0.1 |
GO:1990569 |
UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 |
0.2 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 |
1.2 |
GO:0090102 |
cochlea development(GO:0090102) |
| 0.0 |
0.6 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.0 |
0.1 |
GO:1902035 |
positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 |
0.2 |
GO:0060252 |
positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 |
0.1 |
GO:0010157 |
response to chlorate(GO:0010157) |
| 0.0 |
0.2 |
GO:0044341 |
sodium-dependent phosphate transport(GO:0044341) |
| 0.0 |
0.1 |
GO:0042986 |
positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 |
0.0 |
GO:0060437 |
lung growth(GO:0060437) |
| 0.0 |
0.5 |
GO:0030011 |
maintenance of cell polarity(GO:0030011) |
| 0.0 |
0.0 |
GO:0036333 |
hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 |
0.2 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 |
0.1 |
GO:0034670 |
chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 |
0.2 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.0 |
1.2 |
GO:0010830 |
regulation of myotube differentiation(GO:0010830) |
| 0.0 |
0.1 |
GO:0032927 |
positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 |
0.1 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.5 |
GO:0006198 |
cAMP catabolic process(GO:0006198) |
| 0.0 |
0.1 |
GO:1902498 |
regulation of protein autoubiquitination(GO:1902498) |
| 0.0 |
0.1 |
GO:0072718 |
response to cisplatin(GO:0072718) |
| 0.0 |
0.1 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 |
0.0 |
GO:0006679 |
glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 |
0.1 |
GO:0060013 |
adult feeding behavior(GO:0008343) righting reflex(GO:0060013) |
| 0.0 |
0.1 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.0 |
0.2 |
GO:2001014 |
regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 |
0.1 |
GO:2000324 |
positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 |
0.2 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
0.0 |
GO:0051758 |
homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 |
0.1 |
GO:0007185 |
transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 |
0.0 |
GO:0051083 |
'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 |
0.5 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.7 |
GO:0048384 |
retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 |
0.2 |
GO:0021694 |
cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |