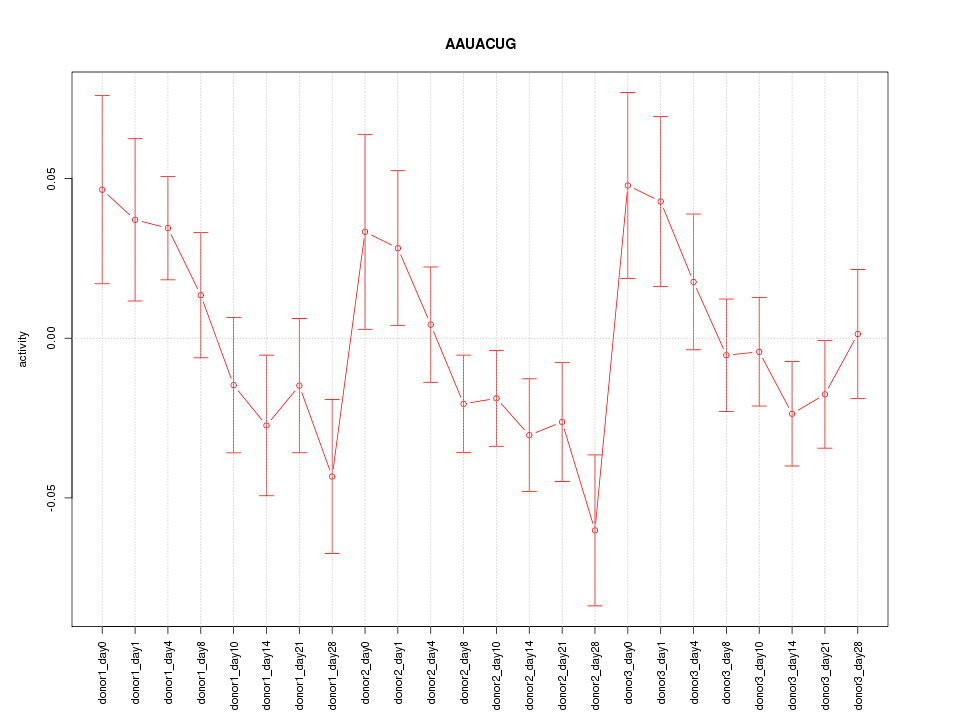
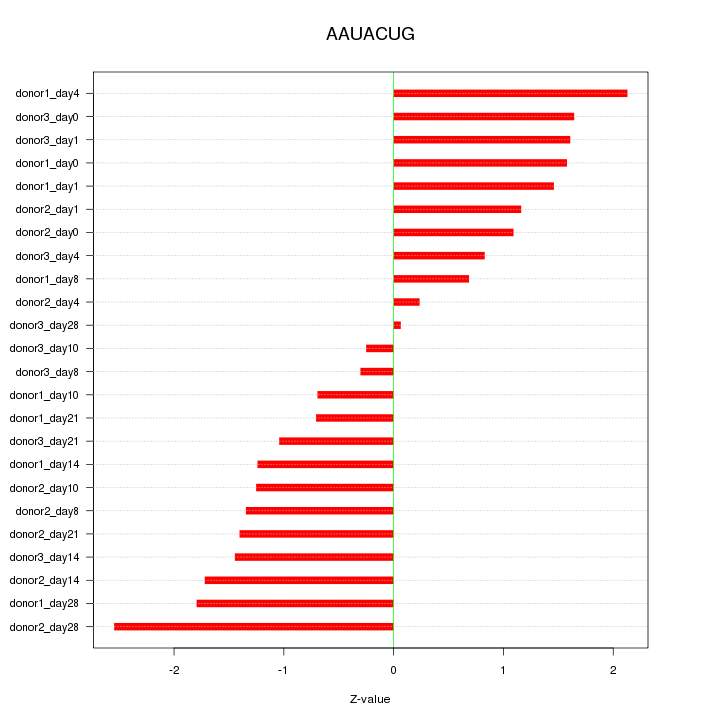
Motif ID: AAUACUG
Z-value: 1.324

Mature miRNA associated with seed AAUACUG:
| Name | miRBase Accession |
|---|---|
| hsa-miR-200b-3p | MIMAT0000318 |
| hsa-miR-200c-3p | MIMAT0000617 |
| hsa-miR-429 | MIMAT0001536 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 1.1 | 3.4 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.8 | 2.5 | GO:1904170 | regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.7 | 4.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.6 | 3.9 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.6 | 1.8 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.6 | 2.3 | GO:0003169 | coronary vein morphogenesis(GO:0003169) |
| 0.5 | 1.9 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.5 | 0.5 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 1.6 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.3 | 1.2 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.3 | 0.9 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.3 | 1.2 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.3 | 1.8 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.3 | 0.9 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.3 | 1.4 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.3 | 0.9 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.3 | 0.8 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.3 | 2.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.3 | 1.9 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.3 | 2.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 0.8 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.3 | 2.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 1.2 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.2 | 1.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 0.9 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 0.9 | GO:0071484 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.2 | 0.7 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.2 | 0.7 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.2 | 1.5 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.2 | 1.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.2 | 0.6 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.2 | 0.6 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.2 | 0.4 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.2 | 4.7 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 2.7 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.2 | 0.8 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.2 | 1.9 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.2 | 1.1 | GO:0030805 | regulation of cyclic nucleotide catabolic process(GO:0030805) regulation of cAMP catabolic process(GO:0030820) regulation of purine nucleotide catabolic process(GO:0033121) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.2 | 0.6 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.2 | 2.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 1.4 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 1.2 | GO:0090625 | siRNA loading onto RISC involved in RNA interference(GO:0035087) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.2 | 0.7 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.2 | 0.7 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.2 | 0.5 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 | 1.0 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.2 | 0.7 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 0.8 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 1.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.2 | 0.8 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.2 | 0.6 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 0.5 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.2 | 0.9 | GO:0051461 | regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.2 | 1.2 | GO:0030578 | PML body organization(GO:0030578) |
| 0.2 | 0.5 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.9 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 2.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 1.6 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.1 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) |
| 0.1 | 0.4 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.8 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.8 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.8 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.1 | 0.4 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.7 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.6 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 0.5 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 1.2 | GO:0015014 | protein sulfation(GO:0006477) heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.4 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.1 | 0.7 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.7 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.2 | GO:0003285 | septum secundum development(GO:0003285) |
| 0.1 | 1.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.2 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 | 0.2 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 0.5 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.1 | 0.4 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.3 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.8 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.1 | 0.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.9 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.6 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.8 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 0.6 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.9 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.6 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 0.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.3 | GO:0015684 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.1 | 0.3 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.5 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.1 | 0.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.3 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.1 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.1 | 0.6 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.4 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.6 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 0.9 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.6 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.4 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.2 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.1 | 0.9 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.2 | GO:2000705 | histone H3-T6 phosphorylation(GO:0035408) dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.5 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.3 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 1.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 1.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.3 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.4 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.1 | 0.3 | GO:0048749 | compound eye development(GO:0048749) |
| 0.1 | 0.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 0.2 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.1 | 0.8 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.9 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.6 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 2.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.7 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.1 | 0.5 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 0.2 | GO:1903347 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 1.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0051462 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) |
| 0.0 | 0.3 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.3 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.6 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.7 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.3 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 1.3 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.1 | GO:0030860 | neuroblast division in subventricular zone(GO:0021849) regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.0 | 0.2 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.6 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.4 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.2 | GO:0046671 | cochlear nucleus development(GO:0021747) negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) regulation of retinal cell programmed cell death(GO:0046668) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.3 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.3 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.9 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 1.4 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.9 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.5 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.7 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 1.0 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 1.4 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.2 | GO:1900453 | negative regulation of long term synaptic depression(GO:1900453) regulation of intracellular transport of viral material(GO:1901252) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.7 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.7 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.7 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.7 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.7 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.0 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.6 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.5 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.4 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.7 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 1.0 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.2 | GO:1902914 | regulation of protein polyubiquitination(GO:1902914) positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 1.1 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 2.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.0 | GO:2001028 | regulation of endothelial cell chemotaxis(GO:2001026) positive regulation of endothelial cell chemotaxis(GO:2001028) |
| 0.0 | 1.9 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 1.7 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) |
| 0.0 | 1.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.9 | GO:0006363 | transcription elongation from RNA polymerase I promoter(GO:0006362) termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.4 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.4 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 1.0 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:1903624 | regulation of apoptotic DNA fragmentation(GO:1902510) regulation of DNA catabolic process(GO:1903624) |
| 0.0 | 0.1 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:1902746 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.2 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.6 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 0.4 | GO:0043536 | positive regulation of blood vessel endothelial cell migration(GO:0043536) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.0 | GO:0071545 | inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.7 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.0 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 2.6 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.0 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.7 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.0 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.1 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.6 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.8 | 4.1 | GO:0044393 | microspike(GO:0044393) |
| 0.7 | 2.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.6 | 2.2 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.5 | 2.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 1.3 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.4 | 1.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.3 | 3.8 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.3 | 0.8 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.3 | 3.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 1.0 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.2 | 0.7 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.2 | 1.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 0.8 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.2 | 0.6 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.2 | 0.9 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.2 | 0.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.6 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 0.9 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 1.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.5 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.1 | 1.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 1.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 0.3 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 1.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.4 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 1.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.4 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 1.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.6 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.6 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 1.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 1.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.4 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.9 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.6 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 2.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.8 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 1.2 | GO:1990752 | microtubule end(GO:1990752) |
| 0.0 | 0.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.9 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.9 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.6 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 1.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 2.6 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 2.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 3.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.6 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.4 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 0.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.7 | 3.7 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.5 | 1.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.4 | 1.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.4 | 0.4 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.3 | 1.0 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.3 | 0.3 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.3 | 2.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 1.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.3 | 1.2 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.3 | 0.9 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.3 | 0.9 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.3 | 0.8 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.3 | 1.3 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.2 | 0.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 1.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 1.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.8 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.2 | 0.8 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.2 | 0.6 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.2 | 0.6 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.2 | 1.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 0.9 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.2 | 2.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 0.7 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 1.3 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 2.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 3.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 3.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.7 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.1 | 0.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.6 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.5 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 0.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.6 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.5 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.1 | 0.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.5 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.9 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.8 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.4 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.2 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 1.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.3 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.2 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 0.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.7 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 2.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.2 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 1.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 3.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 9.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.5 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.3 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 3.8 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 1.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 2.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.6 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 1.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.3 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 3.6 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 2.0 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.3 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.1 | 7.3 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.2 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.0 | SA_TRKA_RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 1.5 | PID_S1P_S1P1_PATHWAY | S1P1 pathway |
| 0.1 | 4.3 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.1 | 1.4 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 2.8 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.1 | 2.2 | PID_EPO_PATHWAY | EPO signaling pathway |
| 0.1 | 2.3 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.4 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.5 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.0 | 2.5 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.4 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.4 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 3.9 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 3.3 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID_ECADHERIN_KERATINOCYTE_PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.2 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.3 | PID_ER_NONGENOMIC_PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.8 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.1 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.2 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.9 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 0.9 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.0 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.1 | PID_VEGFR1_PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.4 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.8 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.3 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | ST_INTERLEUKIN_4_PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.6 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.1 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 0.9 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 0.7 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID_SHP2_PATHWAY | SHP2 signaling |
| 0.0 | 0.5 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.4 | PID_PI3KCI_PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.5 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 1.0 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 3.4 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.4 | REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 4.4 | REACTOME_REGULATION_OF_SIGNALING_BY_CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.8 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.7 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 3.3 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.4 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.9 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.1 | 2.3 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 3.3 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.0 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.0 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.2 | REACTOME_ADENYLATE_CYCLASE_INHIBITORY_PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 1.7 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 3.2 | REACTOME_REGULATION_OF_HYPOXIA_INDUCIBLE_FACTOR_HIF_BY_OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 0.7 | REACTOME_P75NTR_RECRUITS_SIGNALLING_COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 1.2 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.7 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.0 | REACTOME_SIGNALING_BY_CONSTITUTIVELY_ACTIVE_EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.3 | REACTOME_REGULATION_OF_AMPK_ACTIVITY_VIA_LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.7 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.9 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.1 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME_GRB2_SOS_PROVIDES_LINKAGE_TO_MAPK_SIGNALING_FOR_INTERGRINS_ | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.7 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.9 | REACTOME_PHOSPHORYLATION_OF_CD3_AND_TCR_ZETA_CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.7 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.0 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.8 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.0 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.7 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.7 | REACTOME_PERK_REGULATED_GENE_EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.4 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.8 | REACTOME_RNA_POL_II_PRE_TRANSCRIPTION_EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 0.1 | REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.2 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.0 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME_REGULATION_OF_WATER_BALANCE_BY_RENAL_AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.5 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.5 | REACTOME_HYALURONAN_METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.7 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.5 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME_GABA_B_RECEPTOR_ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 1.0 | REACTOME_NITRIC_OXIDE_STIMULATES_GUANYLATE_CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.4 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.0 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME_KERATAN_SULFATE_KERATIN_METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.6 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.4 | REACTOME_PHOSPHORYLATION_OF_THE_APC_C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.1 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.1 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |