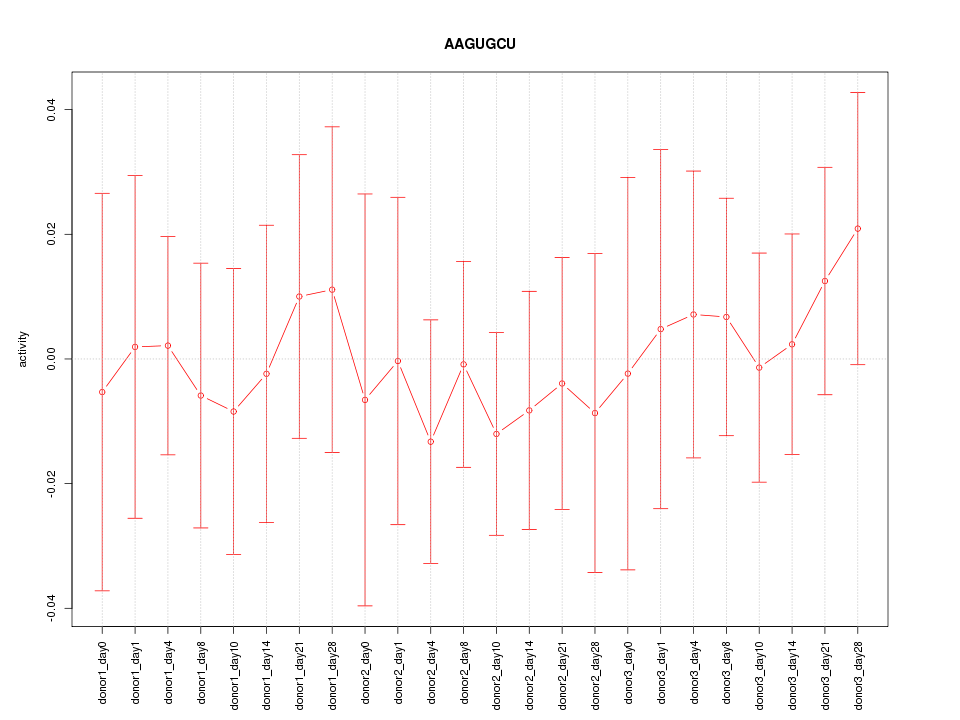
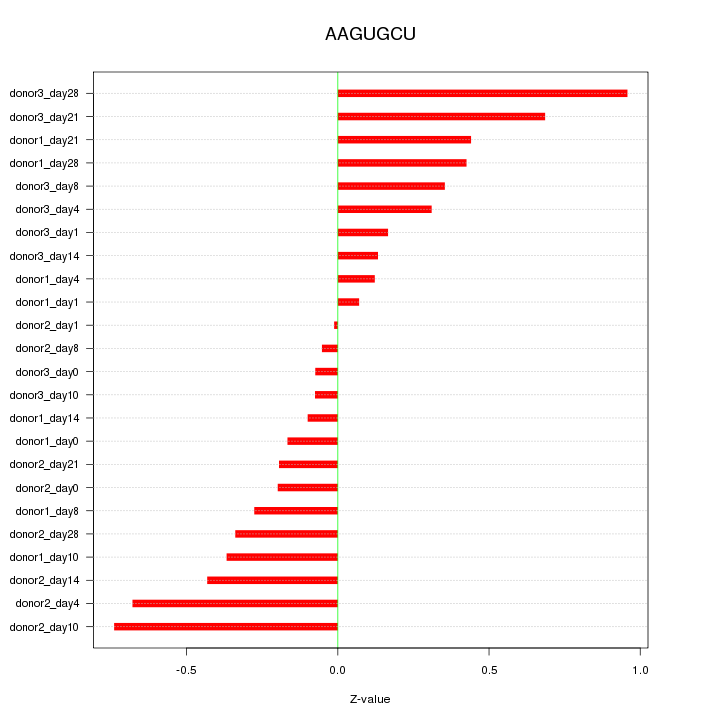
| 0.1 |
0.3 |
GO:0071335 |
proximal/distal axis specification(GO:0009946) bronchiole development(GO:0060435) submandibular salivary gland formation(GO:0060661) hair follicle cell proliferation(GO:0071335) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.1 |
0.3 |
GO:1990086 |
lens fiber cell apoptotic process(GO:1990086) |
| 0.1 |
0.3 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 |
0.3 |
GO:0052551 |
response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 |
0.1 |
GO:0034970 |
histone H3-R2 methylation(GO:0034970) |
| 0.0 |
0.1 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.0 |
0.1 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 |
0.2 |
GO:1903977 |
positive regulation of glial cell migration(GO:1903977) |
| 0.0 |
0.2 |
GO:0071233 |
cellular response to leucine(GO:0071233) |
| 0.0 |
0.1 |
GO:0042264 |
peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 |
0.1 |
GO:0072156 |
distal tubule morphogenesis(GO:0072156) metanephric proximal tubule development(GO:0072237) |
| 0.0 |
0.1 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.0 |
0.1 |
GO:0001555 |
oocyte growth(GO:0001555) positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 |
0.1 |
GO:0070346 |
positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 |
0.1 |
GO:1904760 |
myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 |
0.1 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 |
0.0 |
GO:0070373 |
negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 |
0.1 |
GO:0070839 |
divalent metal ion export(GO:0070839) |
| 0.0 |
0.1 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 |
0.2 |
GO:0032286 |
central nervous system myelin maintenance(GO:0032286) |
| 0.0 |
0.2 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 |
0.1 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 |
0.1 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
0.0 |
GO:0060931 |
sinoatrial node cell development(GO:0060931) |
| 0.0 |
0.1 |
GO:0021831 |
embryonic olfactory bulb interneuron precursor migration(GO:0021831) |
| 0.0 |
0.0 |
GO:0019072 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 |
0.2 |
GO:0021694 |
cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 |
0.0 |
GO:1900224 |
positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 |
0.0 |
GO:0009786 |
regulation of asymmetric cell division(GO:0009786) |
| 0.0 |
0.1 |
GO:0071630 |
nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 |
0.3 |
GO:0071481 |
cellular response to X-ray(GO:0071481) |
| 0.0 |
0.1 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 0.0 |
0.0 |
GO:0002537 |
nitric oxide production involved in inflammatory response(GO:0002537) |
| 0.0 |
0.0 |
GO:1900222 |
regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 |
0.0 |
GO:0045976 |
serotonergic neuron axon guidance(GO:0036515) negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.0 |
0.0 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 |
0.1 |
GO:0038026 |
reelin-mediated signaling pathway(GO:0038026) |
| 0.0 |
0.0 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 |
0.0 |
GO:1990167 |
protein K27-linked deubiquitination(GO:1990167) |
| 0.0 |
0.0 |
GO:1904562 |
phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |